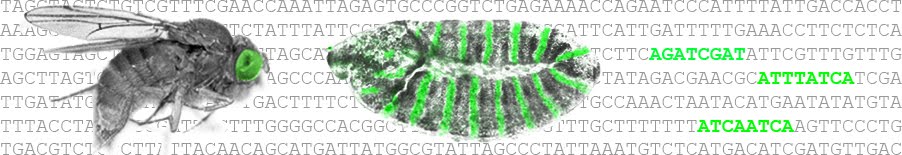
Neural-specific interpretation of morphogens [Developmental Biology]:
The reiterative deployment of a small cadre of morphogen signals underlies patterning and growth of most tissues during embyogenesis, but how such inductive events result in tissue-specific responses remains poorly understood. By characterizing cis-regulatory modules (CRMs) associated with genes regulated by Sonic hedgehog (Shh), retinoids, or bone morphogenetic proteins in...
Tuesday, April 30, 2013
gypsy insulators in a vector mosquito [Applied Biological Sciences]
gypsy insulators in a vector mosquito [Applied Biological Sciences]: Malaria parasites are transmitted to humans by mosquitoes of the genus Anopheles, and these insects are the targets of innovative vector control programs. Proposed approaches include the use of genetic strategies based on transgenic mosquitoes to suppress or modify vector populations. Although substantial advances have been made in engineering resistant...
Predatory cannibalism in Drosophila melanogaster larvae
Predatory cannibalism in Drosophila melanogaster larvae:
Predatory cannibalism in Drosophila melanogaster larvae
Nature Communications 4, 1789 (2013). doi:10.1038/ncomms2744
Authors: Roshan K. Vijendravarma, Sunitha Narasimha & Tadeusz J. Kawecki

Predatory cannibalism in Drosophila melanogaster larvae
Nature Communications 4, 1789 (2013). doi:10.1038/ncomms2744
Authors: Roshan K. Vijendravarma, Sunitha Narasimha & Tadeusz J. Kawecki
Sex-Specific Pattern Formation During Early Drosophila Development [Developmental and Behavioral Genetics]
Sex-Specific Pattern Formation During Early Drosophila Development [Developmental and Behavioral Genetics]:
The deleterious effects of different X-chromosome dosage in males and females are buffered by a process called dosage compensation, which in Drosophila is achieved through a doubling of X-linked transcription in males. The male-specific lethal complex mediates this process, but is known to act only after gastrulation. Recent work has shown that the transcription of X-linked genes is also upregulated in males prior to gastrulation; whether it results in functional dosage compensation is not known. Absent or partial early dosage compensation raises the possibility of sex-biased expression of key developmental genes, such as the segmentation genes controlling anteroposterior patterning. We assess the functional output of early dosage compensation by measuring the expression of even-skipped (eve) with high spatiotemporal resolution in male and female embryos. We show that eve has a sexually dimorphic pattern, suggesting an interaction with either X-chromosome dose or the sex determination system. By manipulating the gene copy number of an X-linked transcription factor, giant (gt), we traced sex-biased eve patterning to gt dose, indicating that early dosage compensation is functionally incomplete. Despite sex-biased eve expression, the gene networks downstream of eve are able to produce sex-independent segmentation, a point that we establish by measuring the proportions of segments in elongated germ-band embryos. Finally, we use a whole-locus eve transgene with modified cis regulation to demonstrate that segment proportions have a sex-dependent sensitivity to subtle changes in Eve expression. The sex independence of downstream segmentation despite this sensitivity to Eve expression implies that additional autosomal gene- or pathway-specific mechanisms are required to ameliorate the effects of partial early dosage compensation.
The deleterious effects of different X-chromosome dosage in males and females are buffered by a process called dosage compensation, which in Drosophila is achieved through a doubling of X-linked transcription in males. The male-specific lethal complex mediates this process, but is known to act only after gastrulation. Recent work has shown that the transcription of X-linked genes is also upregulated in males prior to gastrulation; whether it results in functional dosage compensation is not known. Absent or partial early dosage compensation raises the possibility of sex-biased expression of key developmental genes, such as the segmentation genes controlling anteroposterior patterning. We assess the functional output of early dosage compensation by measuring the expression of even-skipped (eve) with high spatiotemporal resolution in male and female embryos. We show that eve has a sexually dimorphic pattern, suggesting an interaction with either X-chromosome dose or the sex determination system. By manipulating the gene copy number of an X-linked transcription factor, giant (gt), we traced sex-biased eve patterning to gt dose, indicating that early dosage compensation is functionally incomplete. Despite sex-biased eve expression, the gene networks downstream of eve are able to produce sex-independent segmentation, a point that we establish by measuring the proportions of segments in elongated germ-band embryos. Finally, we use a whole-locus eve transgene with modified cis regulation to demonstrate that segment proportions have a sex-dependent sensitivity to subtle changes in Eve expression. The sex independence of downstream segmentation despite this sensitivity to Eve expression implies that additional autosomal gene- or pathway-specific mechanisms are required to ameliorate the effects of partial early dosage compensation.
Mechanisms of tentacle morphogenesis in the sea anemone Nematostella vectensis [RESEARCH ARTICLES]
Mechanisms of tentacle morphogenesis in the sea anemone Nematostella vectensis [RESEARCH ARTICLES]: Ashleigh E. Fritz, Aissam Ikmi, Christopher Seidel, Ariel Paulson, and Matthew C. Gibson
Evolution of the capacity to form secondary outgrowths from the principal embryonic axes was a crucial innovation that potentiated the diversification of animal body plans. Precisely how such outgrowths develop in early-branching metazoan species remains poorly understood. Here we demonstrate that three fundamental processes contribute to embryonic tentacle development in the cnidarian Nematostella vectensis. First, a pseudostratified ectodermal placode forms at the oral pole of developing larvae and is transcriptionally patterned into four tentacle buds. Subsequently, Notch signaling-dependent changes in apicobasal epithelial thickness drive elongation of these primordia. In parallel, oriented cell rearrangements revealed by clonal analysis correlate with shaping of the elongating tentacles. Taken together, our results define the mechanism of embryonic appendage development in an early-branching metazoan, and thereby provide a novel foundation for understanding the diversification of body plans during animal evolution.
Evolution of the capacity to form secondary outgrowths from the principal embryonic axes was a crucial innovation that potentiated the diversification of animal body plans. Precisely how such outgrowths develop in early-branching metazoan species remains poorly understood. Here we demonstrate that three fundamental processes contribute to embryonic tentacle development in the cnidarian Nematostella vectensis. First, a pseudostratified ectodermal placode forms at the oral pole of developing larvae and is transcriptionally patterned into four tentacle buds. Subsequently, Notch signaling-dependent changes in apicobasal epithelial thickness drive elongation of these primordia. In parallel, oriented cell rearrangements revealed by clonal analysis correlate with shaping of the elongating tentacles. Taken together, our results define the mechanism of embryonic appendage development in an early-branching metazoan, and thereby provide a novel foundation for understanding the diversification of body plans during animal evolution.
Bithorax-complex genes sculpt the pattern of leucokinergic neurons in the Drosophila central nervous system [RESEARCH ARTICLES]
Bithorax-complex genes sculpt the pattern of leucokinergic neurons in the Drosophila central nervous system [RESEARCH ARTICLES]: Alicia Estacio-Gomez, Marta Moris-Sanz, Anne-Kathrin Schafer, Daniel Perea, Pilar Herrero, and Fernando J. Diaz-Benjumea
Although the Hox genes are the main factors involved in the generation of diversity along the anterior/posterior body axis of segmented organisms, it is still largely unknown how these genes act in single cells to determine specific traits at precise developmental stages. The aim of this study was to understand the mechanisms by which Hox genes of the Bithorax complex (Bx-C) of Drosophila act to define segmental differences in the ventral nerve cord of the central nervous system. To achieve this, we have focused on the specification of the leucokinin-expressing neurons. We find that these neurons are specified from the same progenitor neuroblast at two different developmental stages: embryonic and larval neurogenesis. We show that genes of the Bx-C acted in postmitotic cells to specify the segment-specific appearance of leucokinergic cells in the larval and adult ventral nerve cord.
Although the Hox genes are the main factors involved in the generation of diversity along the anterior/posterior body axis of segmented organisms, it is still largely unknown how these genes act in single cells to determine specific traits at precise developmental stages. The aim of this study was to understand the mechanisms by which Hox genes of the Bithorax complex (Bx-C) of Drosophila act to define segmental differences in the ventral nerve cord of the central nervous system. To achieve this, we have focused on the specification of the leucokinin-expressing neurons. We find that these neurons are specified from the same progenitor neuroblast at two different developmental stages: embryonic and larval neurogenesis. We show that genes of the Bx-C acted in postmitotic cells to specify the segment-specific appearance of leucokinergic cells in the larval and adult ventral nerve cord.
Decoupling the function of Hox and Shh in developing limb reveals multiple inputs of Hox genes on limb growth [RESEARCH ARTICLES]
Decoupling the function of Hox and Shh in developing limb reveals multiple inputs of Hox genes on limb growth [RESEARCH ARTICLES]: Rushikesh Sheth, Damien Gregoire, Annie Dumouchel, Martina Scotti, Jessica My Trang Pham, Stephen Nemec, Maria Felix Bastida, Marian A. Ros, and Marie Kmita
Limb development relies on an exquisite coordination between growth and patterning, but the underlying mechanisms remain elusive. Anterior-posterior and proximal-distal specification initiates in early limb bud concomitantly with the proliferative expansion of limb cells. Previous studies have shown that limb bud growth initially relies on fibroblast growth factors (FGFs) produced in the apical ectodermal ridge (AER-FGFs), the maintenance of which relies on a positive-feedback loop involving sonic hedgehog (Shh) and the BMP antagonist gremlin 1 (Grem1). The positive cross-regulation between Shh and the HoxA and HoxD clustered genes identified an indirect effect of Hox genes on the maintenance of AER-FGFs but the respective function of Shh and Hox genes in this process remains unknown. Here, by uncoupling Hox and Shh function, we show that HoxA and HoxD genes are required for proper AER-FGFs expression, independently of their function in controlling Shh expression. In addition, we provide evidence that the Hox-dependent control of AER-FGF expression is achieved through the regulation of key mesenchymal signals, namely Grem1 and Fgf10, ensuring proper epithelial-mesenchymal interactions. Notably, HoxA and HoxD genes contribute to both the initial activation of Grem1 and the subsequent anterior expansion of its expression domain. We propose that the intricate interactions between Hox genes and the FGF and Shh signaling pathways act as a molecular network that ensures proper limb bud growth and patterning, probably contributing to the coordination of these two processes.
Limb development relies on an exquisite coordination between growth and patterning, but the underlying mechanisms remain elusive. Anterior-posterior and proximal-distal specification initiates in early limb bud concomitantly with the proliferative expansion of limb cells. Previous studies have shown that limb bud growth initially relies on fibroblast growth factors (FGFs) produced in the apical ectodermal ridge (AER-FGFs), the maintenance of which relies on a positive-feedback loop involving sonic hedgehog (Shh) and the BMP antagonist gremlin 1 (Grem1). The positive cross-regulation between Shh and the HoxA and HoxD clustered genes identified an indirect effect of Hox genes on the maintenance of AER-FGFs but the respective function of Shh and Hox genes in this process remains unknown. Here, by uncoupling Hox and Shh function, we show that HoxA and HoxD genes are required for proper AER-FGFs expression, independently of their function in controlling Shh expression. In addition, we provide evidence that the Hox-dependent control of AER-FGF expression is achieved through the regulation of key mesenchymal signals, namely Grem1 and Fgf10, ensuring proper epithelial-mesenchymal interactions. Notably, HoxA and HoxD genes contribute to both the initial activation of Grem1 and the subsequent anterior expansion of its expression domain. We propose that the intricate interactions between Hox genes and the FGF and Shh signaling pathways act as a molecular network that ensures proper limb bud growth and patterning, probably contributing to the coordination of these two processes.
Monday, April 29, 2013
Single-molecule imaging of transcription factor binding to DNA in live mammalian cells
Single-molecule imaging of transcription factor binding to DNA in live mammalian cells:
Nature Methods 10, 421 (2013).
doi:10.1038/nmeth.2411
Authors: J Christof M Gebhardt, David M Suter, Rahul Roy, Ziqing W Zhao, Alec R Chapman, Srinjan Basu, Tom Maniatis & X Sunney Xie

Nature Methods 10, 421 (2013).
doi:10.1038/nmeth.2411
Authors: J Christof M Gebhardt, David M Suter, Rahul Roy, Ziqing W Zhao, Alec R Chapman, Srinjan Basu, Tom Maniatis & X Sunney Xie
Friday, April 26, 2013
PUPARIATION SITE PREFERENCE WITHIN AND BETWEEN DROSOPHILA SIBLING SPECIES
PUPARIATION SITE PREFERENCE WITHIN AND BETWEEN DROSOPHILA SIBLING SPECIES:
Abstract
Holometabolous insects pass through a sedentary pupal stage and often choose a location for pupation that is different from the site of larval feeding. We have characterized a difference in pupariation site choice within and between sibling species of Drosophila. We found that, in nature, D. sechellia pupariate within their host fruit, Morinda citrifolia, and that they perform this behavior in laboratory assays. In contrast, in the laboratory, geographically diverse strains of D. simulans vary in their pupariation site preference; D. simulans lines from the ancestral range in southeast Africa pupariate on fruit, or a fruit substitute, while populations from Europe or the New World select sites off of fruit. We explored the genetic basis for the evolved preference in puariation site preference by performing quantitative trait locus (QTL) mapping within and between species. We found that the inter-specific difference is controlled largely by loci on chromosomes X and II. In contrast, variation between two strains of D. simulans appears to be highly polygenic, with the majority of phenotypic effects due to loci on chromosome III. These data address the genetic basis of how new traits arise as species diverge and populations disperse.
This article is protected by copyright. All rights reserved.
Linking Genomics and Ecology to Investigate the Complex Evolution of an Invasive Drosophila Pest
Linking Genomics and Ecology to Investigate the Complex Evolution of an Invasive Drosophila Pest:
Drosophilid fruit flies have provided science with striking cases of behavioral adaptation and genetic innovation. A recent example is the invasive pest Drosophila suzukii, which, unlike most other Drosophila, lays eggs and feeds on undamaged, ripening fruits. This not only poses a serious threat for fruit cultivation but also offers an interesting model to study evolution of behavioral innovation. We developed genome and transcriptome resources for D. suzukii. Coupling analyses of these data with field observations, we propose a hypothesis of the origin of its peculiar ecology. Using nuclear and mitochondrial phylogenetic analyses, we confirm its Asian origin and reveal a surprising sister relationship between the eugracilis and the melanogaster subgroups. Although the D. suzukii genome is comparable in size and repeat content to other Drosophila species, it has the lowest nucleotide substitution rate among the species analyzed in this study. This finding is compatible with the overwintering diapause of D. suzukii, which results in a reduced number of generations per year compared with its sister species. Genome-scale relaxed clock analyses support a late Miocene origin of D. suzukii, concomitant with paleogeological and climatic conditions that suggest an adaptation to temperate montane forests, a hypothesis confirmed by field trapping. We propose a causal link between the ecological adaptations of D. suzukii in its native habitat and its invasive success in Europe and North America.
Drosophilid fruit flies have provided science with striking cases of behavioral adaptation and genetic innovation. A recent example is the invasive pest Drosophila suzukii, which, unlike most other Drosophila, lays eggs and feeds on undamaged, ripening fruits. This not only poses a serious threat for fruit cultivation but also offers an interesting model to study evolution of behavioral innovation. We developed genome and transcriptome resources for D. suzukii. Coupling analyses of these data with field observations, we propose a hypothesis of the origin of its peculiar ecology. Using nuclear and mitochondrial phylogenetic analyses, we confirm its Asian origin and reveal a surprising sister relationship between the eugracilis and the melanogaster subgroups. Although the D. suzukii genome is comparable in size and repeat content to other Drosophila species, it has the lowest nucleotide substitution rate among the species analyzed in this study. This finding is compatible with the overwintering diapause of D. suzukii, which results in a reduced number of generations per year compared with its sister species. Genome-scale relaxed clock analyses support a late Miocene origin of D. suzukii, concomitant with paleogeological and climatic conditions that suggest an adaptation to temperate montane forests, a hypothesis confirmed by field trapping. We propose a causal link between the ecological adaptations of D. suzukii in its native habitat and its invasive success in Europe and North America.
Germ Cell Specification Requires Zygotic Mechanisms Rather Than Germ Plasm in a Basally Branching Insect
Germ Cell Specification Requires Zygotic Mechanisms Rather Than Germ Plasm in a Basally Branching Insect: Ben Ewen-Campen, Seth Donoughe, Donald Nat Clarke, Cassandra G. Extavour. BackgroundPrimordial germ cell (PGC) specification is a universal process across animals, but the molecular mechanisms specifying PGCs are remarkably diverse. In Drosophila, PGCs are specif....
Thursday, April 25, 2013
Specified Neural Progenitors Sort to Form Sharp Domains after Noisy Shh Signaling
Specified Neural Progenitors Sort to Form Sharp Domains after Noisy Shh Signaling: Fengzhu Xiong, Andrea R. Tentner, Peng Huang, Arnaud Gelas, Kishore R. Mosaliganti, Lydie Souhait, Nicolas Rannou, Ian A. Swinburne, Nikolaus D. Obholzer, Paul D. Cowgill, Alexander F. Schier, Sean G. Megason. Sharply delineated domains of cell types arise in developing tissues under instruction of inductive signal (morphogen) gradients, which specify distinct cell fates at different signal levels. The ....
1953: When Genes Became “Information”
1953: When Genes Became “Information”: Matthew Cobb. In 1953, Watson and Crick not only described the double-helix structure of DNA, but also embraced the idea that genes contained a code that expresses information and thereby changed our view of li....
[Report] Insect Morphological Diversification Through the Modification of Wing Serial Homologs
[Report] Insect Morphological Diversification Through the Modification of Wing Serial Homologs: Modification rather than loss of dorsal appendages has provided a diversifying mechanism for the insect body plan.
Authors: Takahiro Ohde, Toshinobu Yaginuma, Teruyuki Niimi
Authors: Takahiro Ohde, Toshinobu Yaginuma, Teruyuki Niimi
Six Homeoproteins Directly Activate Myod Expression in the Gene Regulatory Networks That Control Early Myogenesis
Six Homeoproteins Directly Activate Myod Expression in the Gene Regulatory Networks That Control Early Myogenesis:
by Frédéric Relaix, Josiane Demignon, Christine Laclef, Julien Pujol, Marc Santolini, Claire Niro, Mounia Lagha, Didier Rocancourt, Margaret Buckingham, Pascal Maire
In mammals, several genetic pathways have been characterized that govern engagement of multipotent embryonic progenitors into the myogenic program through the control of the key myogenic regulatory gene Myod. Here we demonstrate the involvement of Six homeoproteins. We first targeted into a Pax3 allele a sequence encoding a negative form of Six4 that binds DNA but cannot interact with essential Eya co-factors. The resulting embryos present hypoplasic skeletal muscles and impaired Myod activation in the trunk in the absence of Myf5/Mrf4. At the axial level, we further show that Myod is still expressed in compound Six1/Six4:Pax3 but not in Six1/Six4:Myf5 triple mutant embryos, demonstrating that Six1/4 participates in the Pax3-Myod genetic pathway. Myod expression and head myogenesis is preserved in Six1/Six4:Myf5 triple mutant embryos, illustrating that upstream regulators of Myod in different embryonic territories are distinct. We show that Myod regulatory regions are directly controlled by Six proteins and that, in the absence of Six1 and Six4, Six2 can compensate.
by Frédéric Relaix, Josiane Demignon, Christine Laclef, Julien Pujol, Marc Santolini, Claire Niro, Mounia Lagha, Didier Rocancourt, Margaret Buckingham, Pascal Maire
In mammals, several genetic pathways have been characterized that govern engagement of multipotent embryonic progenitors into the myogenic program through the control of the key myogenic regulatory gene Myod. Here we demonstrate the involvement of Six homeoproteins. We first targeted into a Pax3 allele a sequence encoding a negative form of Six4 that binds DNA but cannot interact with essential Eya co-factors. The resulting embryos present hypoplasic skeletal muscles and impaired Myod activation in the trunk in the absence of Myf5/Mrf4. At the axial level, we further show that Myod is still expressed in compound Six1/Six4:Pax3 but not in Six1/Six4:Myf5 triple mutant embryos, demonstrating that Six1/4 participates in the Pax3-Myod genetic pathway. Myod expression and head myogenesis is preserved in Six1/Six4:Myf5 triple mutant embryos, illustrating that upstream regulators of Myod in different embryonic territories are distinct. We show that Myod regulatory regions are directly controlled by Six proteins and that, in the absence of Six1 and Six4, Six2 can compensate.
Wednesday, April 24, 2013
Dynamic regulatory network controlling TH17 cell differentiation
Dynamic regulatory network controlling TH17 cell differentiation:
Dynamic regulatory network controlling TH17 cell differentiation
Nature 496, 7446 (2013). doi:10.1038/nature11981
Authors: Nir Yosef, Alex K. Shalek, Jellert T. Gaublomme, Hulin Jin, Youjin Lee, Amit Awasthi, Chuan Wu, Katarzyna Karwacz, Sheng Xiao, Marsela Jorgolli, David Gennert, Rahul Satija, Arvind Shakya, Diana Y. Lu, John J. Trombetta, Meenu R. Pillai, Peter J. Ratcliffe, Mathew L. Coleman, Mark Bix, Dean Tantin, Hongkun Park, Vijay K. Kuchroo & Aviv Regev
Despite their importance, the molecular circuits that control the differentiation of naive T cells remain largely unknown. Recent studies that reconstructed regulatory networks in mammalian cells have focused on short-term responses and relied on perturbation-based approaches that cannot be readily applied to primary T cells.

Dynamic regulatory network controlling TH17 cell differentiation
Nature 496, 7446 (2013). doi:10.1038/nature11981
Authors: Nir Yosef, Alex K. Shalek, Jellert T. Gaublomme, Hulin Jin, Youjin Lee, Amit Awasthi, Chuan Wu, Katarzyna Karwacz, Sheng Xiao, Marsela Jorgolli, David Gennert, Rahul Satija, Arvind Shakya, Diana Y. Lu, John J. Trombetta, Meenu R. Pillai, Peter J. Ratcliffe, Mathew L. Coleman, Mark Bix, Dean Tantin, Hongkun Park, Vijay K. Kuchroo & Aviv Regev
Despite their importance, the molecular circuits that control the differentiation of naive T cells remain largely unknown. Recent studies that reconstructed regulatory networks in mammalian cells have focused on short-term responses and relied on perturbation-based approaches that cannot be readily applied to primary T cells.
Tuesday, April 23, 2013
Quantitative relationships between histone marks [Genetics]
Quantitative relationships between histone marks [Genetics]: Gene expression is controlled by coordinated action of many epigenetic mechanisms including covalent histone modifications. Although numerous recurrent patterns of colocalized histone modifications have been associated with specific gene expression states, interrelationships between individual modifications are largely unknown. Here, we analyze quantitative relationships between colocalized histone marks during embryonic stem...
Dynamic interpretation of input concentration [Developmental Biology]
Dynamic interpretation of input concentration [Developmental Biology]: Patterning of body parts in multicellular organisms relies on the interpretation of transcription factor (TF) concentrations by genetic networks. To determine the extent by which absolute TF concentration dictates gene expression and morphogenesis programs that ultimately lead to patterns in Drosophila embryos, we manipulate maternally supplied patterning determinants and measure...
Proteomic profiling of transcription factors [Biochemistry]
Proteomic profiling of transcription factors [Biochemistry]: Transcription factors (TFs) are families of proteins that bind to specific DNA sequences, or TF response elements (TFREs), and function as regulators of many cellular processes. Because of the low abundance of TFs, direct quantitative measurement of TFs on a proteome scale remains a challenge. In this study, we report...
An Essential Role for Zygotic Expression in the Pre-Cellular Drosophila Embryo
An Essential Role for Zygotic Expression in the Pre-Cellular Drosophila Embryo:
by Zehra Ali-Murthy, Susan E. Lott, Michael B. Eisen, Thomas B. Kornberg
The Drosophila embryo proceeds through thirteen mitotic divisions as a syncytium. Its nuclei distribute in the embryo's interior during the first six divisions, dividing synchronously with a cycle time of less than ten minutes. After seven divisions (nuclear cycle 8), the syncytial blastoderm forms as the nuclei approach the embryo surface and slow their cycle time; subsequent divisions proceed in waves that initiate at the poles. Because genetic studies have not identified zygotic mutants that affect the early divisions and because transcription has not been detected before cycle 8, the early, pre-blastoderm embryo has been considered to rely entirely on maternal contributions and to be transcriptionally silent. Our studies identified several abnormal phenotypes in live engrailed (en) mutant embryos prior to cycle 8, as well as a small group of genes that are transcribed in embryos prior to cycle 7. Nuclei in en embryos divide asynchronously, an abnormality that was detected as early as nuclear cycle 2–3. Anti-En antibody detected nuclear En protein in embryos at cycle 2, and expression of an En:GFP fusion protein encoded in the paternal genome was also detected in cycle 2 nuclei. These findings demonstrate that the Drosophila embryo is functionally competent for gene expression prior to the onset of its rapid nuclear divisions and that the embryo requires functions that are expressed in the zygote in order to faithfully prosecute its early, pre-cellularization mitotic cycles.
by Zehra Ali-Murthy, Susan E. Lott, Michael B. Eisen, Thomas B. Kornberg
The Drosophila embryo proceeds through thirteen mitotic divisions as a syncytium. Its nuclei distribute in the embryo's interior during the first six divisions, dividing synchronously with a cycle time of less than ten minutes. After seven divisions (nuclear cycle 8), the syncytial blastoderm forms as the nuclei approach the embryo surface and slow their cycle time; subsequent divisions proceed in waves that initiate at the poles. Because genetic studies have not identified zygotic mutants that affect the early divisions and because transcription has not been detected before cycle 8, the early, pre-blastoderm embryo has been considered to rely entirely on maternal contributions and to be transcriptionally silent. Our studies identified several abnormal phenotypes in live engrailed (en) mutant embryos prior to cycle 8, as well as a small group of genes that are transcribed in embryos prior to cycle 7. Nuclei in en embryos divide asynchronously, an abnormality that was detected as early as nuclear cycle 2–3. Anti-En antibody detected nuclear En protein in embryos at cycle 2, and expression of an En:GFP fusion protein encoded in the paternal genome was also detected in cycle 2 nuclei. These findings demonstrate that the Drosophila embryo is functionally competent for gene expression prior to the onset of its rapid nuclear divisions and that the embryo requires functions that are expressed in the zygote in order to faithfully prosecute its early, pre-cellularization mitotic cycles.
Thursday, April 18, 2013
Insulators Target Active Genes to Transcription Factories and Polycomb-Repressed Genes to Polycomb Bodies
Insulators Target Active Genes to Transcription Factories and Polycomb-Repressed Genes to Polycomb Bodies:
by Hua-Bing Li, Katsuhito Ohno, Hongxing Gui, Vincenzo Pirrotta
Polycomb bodies are foci of Polycomb proteins in which different Polycomb target genes are thought to co-localize in the nucleus, looping out from their chromosomal context. We have shown previously that insulators, not Polycomb response elements (PREs), mediate associations among Polycomb Group (PcG) targets to form Polycomb bodies. Here we use live imaging and 3C interactions to show that transgenes containing PREs and endogenous PcG-regulated genes are targeted by insulator proteins to different nuclear structures depending on their state of activity. When two genes are repressed, they co-localize in Polycomb bodies. When both are active, they are targeted to transcription factories in a fashion dependent on Trithorax and enhancer specificity as well as the insulator protein CTCF. In the absence of CTCF, assembly of Polycomb bodies is essentially reduced to those representing genomic clusters of Polycomb target genes. The critical role of Trithorax suggests that stable association with a specialized transcription factory underlies the cellular memory of the active state.
by Hua-Bing Li, Katsuhito Ohno, Hongxing Gui, Vincenzo Pirrotta
Polycomb bodies are foci of Polycomb proteins in which different Polycomb target genes are thought to co-localize in the nucleus, looping out from their chromosomal context. We have shown previously that insulators, not Polycomb response elements (PREs), mediate associations among Polycomb Group (PcG) targets to form Polycomb bodies. Here we use live imaging and 3C interactions to show that transgenes containing PREs and endogenous PcG-regulated genes are targeted by insulator proteins to different nuclear structures depending on their state of activity. When two genes are repressed, they co-localize in Polycomb bodies. When both are active, they are targeted to transcription factories in a fashion dependent on Trithorax and enhancer specificity as well as the insulator protein CTCF. In the absence of CTCF, assembly of Polycomb bodies is essentially reduced to those representing genomic clusters of Polycomb target genes. The critical role of Trithorax suggests that stable association with a specialized transcription factory underlies the cellular memory of the active state.
TIP48/Reptin and H2A.Z Requirement for Initiating Chromatin Remodeling in Estrogen-Activated Transcription
TIP48/Reptin and H2A.Z Requirement for Initiating Chromatin Remodeling in Estrogen-Activated Transcription:
by Mathieu Dalvai, Laurence Fleury, Luca Bellucci, Silvia Kocanova, Kerstin Bystricky
Histone variants, including histone H2A.Z, are incorporated into specific genomic sites and participate in transcription regulation. The role of H2A.Z at these sites remains poorly characterized. Our study investigates changes in the chromatin environment at the Cyclin D1 gene (CCND1) during transcriptional initiation in response to estradiol in estrogen receptor positive mammary tumour cells. We show that H2A.Z is present at the transcription start-site and downstream enhancer sequences of CCND1 when the gene is poorly transcribed. Stimulation of CCND1 expression required release of H2A.Z concomitantly from both these DNA elements. The AAA+ family members TIP48/reptin and the histone variant H2A.Z are required to remodel the chromatin environment at CCND1 as a prerequisite for binding of the estrogen receptor (ERα) in the presence of hormone. TIP48 promotes acetylation and exchange of H2A.Z, which triggers a dissociation of the CCND1 3′ enhancer from the promoter, thereby releasing a repressive intragenic loop. This release then enables the estrogen receptor to bind to the CCND1 promoter. Our findings provide new insight into the priming of chromatin required for transcription factor access to their target sequence. Dynamic release of gene loops could be a rapid means to remodel chromatin and to stimulate transcription in response to hormones.
by Mathieu Dalvai, Laurence Fleury, Luca Bellucci, Silvia Kocanova, Kerstin Bystricky
Histone variants, including histone H2A.Z, are incorporated into specific genomic sites and participate in transcription regulation. The role of H2A.Z at these sites remains poorly characterized. Our study investigates changes in the chromatin environment at the Cyclin D1 gene (CCND1) during transcriptional initiation in response to estradiol in estrogen receptor positive mammary tumour cells. We show that H2A.Z is present at the transcription start-site and downstream enhancer sequences of CCND1 when the gene is poorly transcribed. Stimulation of CCND1 expression required release of H2A.Z concomitantly from both these DNA elements. The AAA+ family members TIP48/reptin and the histone variant H2A.Z are required to remodel the chromatin environment at CCND1 as a prerequisite for binding of the estrogen receptor (ERα) in the presence of hormone. TIP48 promotes acetylation and exchange of H2A.Z, which triggers a dissociation of the CCND1 3′ enhancer from the promoter, thereby releasing a repressive intragenic loop. This release then enables the estrogen receptor to bind to the CCND1 promoter. Our findings provide new insight into the priming of chromatin required for transcription factor access to their target sequence. Dynamic release of gene loops could be a rapid means to remodel chromatin and to stimulate transcription in response to hormones.
Population genomics: Characterizing indels
Population genomics: Characterizing indels:
Nature Reviews Genetics 14, 302 (2013).
doi:10.1038/nrg3487
Author: Hannah Stower
Small insertions and deletions (indels) are some of the least well-characterized and least understood variants in the human genome. In this paper, the authors sequenced the genomes of 179 individuals from three populations and identified 1.6 million indels. The authors showed heterogenous rates of indel

Nature Reviews Genetics 14, 302 (2013).
doi:10.1038/nrg3487
Author: Hannah Stower
Small insertions and deletions (indels) are some of the least well-characterized and least understood variants in the human genome. In this paper, the authors sequenced the genomes of 179 individuals from three populations and identified 1.6 million indels. The authors showed heterogenous rates of indel
Wednesday, April 17, 2013
Selective Inhibition of Tumor Oncogenes by Disruption of Super-Enhancers
Selective Inhibition of Tumor Oncogenes by Disruption of Super-Enhancers: Jakob Lovén, Heather A. Hoke, Charles Y. Lin, Ashley Lau, David A. Orlando, Christopher R. Vakoc, James E. Bradner, Tong Ihn Lee, Richard A. Young. Chromatin regulators have become attractive targets for cancer therapy, but it is unclear why inhibition of these ubiquitous regulators should have gene-specific effects in tumor cells. Here, we i....
Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes
Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes: Warren A. Whyte, David A. Orlando, Denes Hnisz, Brian J. Abraham, Charles Y. Lin, Michael H. Kagey, Peter B. Rahl, Tong Ihn Lee, Richard A. Young. Master transcription factors Oct4, Sox2, and Nanog bind enhancer elements and recruit Mediator to activate much of the gene expression program of pluripotent embryonic stem cells (ESCs). We report....
Clonal Development and Organization of the Adult Drosophila Central Brain
Clonal Development and Organization of the Adult Drosophila Central Brain: Hung-Hsiang Yu, Takeshi Awasaki, Mark David Schroeder, Fuhui Long, Jacob S. Yang, Yisheng He, Peng Ding, Jui-Chun Kao, Gloria Yueh-Yi Wu, Hanchuan Peng, Gene Myers, Tzumin Lee. BackgroundThe insect brain can be divided into neuropils that are formed by neurites of both local and remote origin. The complexity of the interconnections obscures how these neuropils are establ....
Variation in the Dorsal Gradient Distribution Is a Source for Modified Scaling of Germ Layers in Drosophila
Variation in the Dorsal Gradient Distribution Is a Source for Modified Scaling of Germ Layers in Drosophila: Juan Sebastian Chahda, Rui Sousa-Neves, Claudia Mieko Mizutani.
Specification of germ layers along the dorsoventral axis by morphogenetic gradients is an ideal model to study scaling properties of gradients and cell fate changes during evolution. Classical ana....
Specification of germ layers along the dorsoventral axis by morphogenetic gradients is an ideal model to study scaling properties of gradients and cell fate changes during evolution. Classical ana....
Visualization of an endogenous retinoic acid gradient across embryonic development
Visualization of an endogenous retinoic acid gradient across embryonic development:
Visualization of an endogenous retinoic acid gradient across embryonic development
Nature 496, 7445 (2013). doi:10.1038/nature12037
Authors: Satoshi Shimozono, Tadahiro Iimura, Tetsuya Kitaguchi, Shin-ichi Higashijima & Atsushi Miyawaki
In vertebrate development, the body plan is determined by primordial morphogen gradients that suffuse the embryo. Retinoic acid (RA) is an important morphogen involved in patterning the anterior–posterior axis of structures, including the hindbrainand paraxial mesoderm. RA diffuses over long distances, and its activity is spatially restricted by synthesizing and degrading enzymes. However, gradients of endogenous morphogens in live embryos have not been directly observed; indeed, their existence, distribution and requirement for correct patterning remain controversial. Here we report a family of genetically encoded indicators for RA that we have termed GEPRAs (genetically encoded probes for RA). Using the principle of fluorescence resonance energy transfer we engineered the ligand-binding domains of RA receptors to incorporate cyan-emitting and yellow-emitting fluorescent proteins as fluorescence resonance energy transfer donor and acceptor, respectively, for the reliable detection of ambient free RA. We created three GEPRAs with different affinities for RA, enabling the quantitative measurement of physiological RA concentrations. Live imaging of zebrafish embryos at the gastrula and somitogenesis stages revealed a linear concentration gradient of endogenous RA in a two-tailed source–sink arrangement across the embryo. Modelling of the observed linear RA gradient suggests that the rate of RA diffusion exceeds the spatiotemporal dynamics of embryogenesis, resulting in stability to perturbation. Furthermore, we used GEPRAs in combination with genetic and pharmacological perturbations to resolve competing hypotheses on the structure of the RA gradient during hindbrain formation and somitogenesis. Live imaging of endogenous concentration gradients across embryonic development will allow the precise assignment of molecular mechanisms to developmental dynamics and will accelerate the application of approaches based on morphogen gradients to tissue engineering and regenerative medicine.

Visualization of an endogenous retinoic acid gradient across embryonic development
Nature 496, 7445 (2013). doi:10.1038/nature12037
Authors: Satoshi Shimozono, Tadahiro Iimura, Tetsuya Kitaguchi, Shin-ichi Higashijima & Atsushi Miyawaki
In vertebrate development, the body plan is determined by primordial morphogen gradients that suffuse the embryo. Retinoic acid (RA) is an important morphogen involved in patterning the anterior–posterior axis of structures, including the hindbrainand paraxial mesoderm. RA diffuses over long distances, and its activity is spatially restricted by synthesizing and degrading enzymes. However, gradients of endogenous morphogens in live embryos have not been directly observed; indeed, their existence, distribution and requirement for correct patterning remain controversial. Here we report a family of genetically encoded indicators for RA that we have termed GEPRAs (genetically encoded probes for RA). Using the principle of fluorescence resonance energy transfer we engineered the ligand-binding domains of RA receptors to incorporate cyan-emitting and yellow-emitting fluorescent proteins as fluorescence resonance energy transfer donor and acceptor, respectively, for the reliable detection of ambient free RA. We created three GEPRAs with different affinities for RA, enabling the quantitative measurement of physiological RA concentrations. Live imaging of zebrafish embryos at the gastrula and somitogenesis stages revealed a linear concentration gradient of endogenous RA in a two-tailed source–sink arrangement across the embryo. Modelling of the observed linear RA gradient suggests that the rate of RA diffusion exceeds the spatiotemporal dynamics of embryogenesis, resulting in stability to perturbation. Furthermore, we used GEPRAs in combination with genetic and pharmacological perturbations to resolve competing hypotheses on the structure of the RA gradient during hindbrain formation and somitogenesis. Live imaging of endogenous concentration gradients across embryonic development will allow the precise assignment of molecular mechanisms to developmental dynamics and will accelerate the application of approaches based on morphogen gradients to tissue engineering and regenerative medicine.
Tuesday, April 16, 2013
In vivo live imaging of RNA polymerase II transcription factories in primary cells [Research Papers]
In vivo live imaging of RNA polymerase II transcription factories in primary cells [Research Papers]:
Transcription steps are marked by different modifications of the C-terminal domain of RNA polymerase II (RNAPII). Phosphorylation of Ser5 and Ser7 by cyclin-dependent kinase 7 (CDK7) as part of TFIIH marks initiation, whereas phosphorylation of Ser2 by CDK9 marks elongation. These processes are thought to take place in localized transcription foci in the nucleus, known as "transcription factories," but it has been argued that the observed clusters/foci are mere fixation or labeling artifacts. We show that transcription factories exist in living cells as distinct foci by live-imaging fluorescently labeled CDK9, a kinase known to associate with active RNAPII. These foci were observed in different cell types derived from CDK9-mCherry knock-in mice. We show that these foci are very stable while highly dynamic in exchanging CDK9. Chromatin immunoprecipitation (ChIP) coupled with deep sequencing (ChIP-seq) data show that the genome-wide binding sites of CDK9 and initiating RNAPII overlap on transcribed genes. Immunostaining shows that CDK9-mCherry foci colocalize with RNAPII-Ser5P, much less with RNAPII-Ser2P, and not with CDK12 (a kinase reported to be involved in the Ser2 phosphorylation) or with splicing factor SC35. In conclusion, transcription factories exist in living cells, and initiation and elongation of transcripts takes place in different nuclear compartments.
Transcription steps are marked by different modifications of the C-terminal domain of RNA polymerase II (RNAPII). Phosphorylation of Ser5 and Ser7 by cyclin-dependent kinase 7 (CDK7) as part of TFIIH marks initiation, whereas phosphorylation of Ser2 by CDK9 marks elongation. These processes are thought to take place in localized transcription foci in the nucleus, known as "transcription factories," but it has been argued that the observed clusters/foci are mere fixation or labeling artifacts. We show that transcription factories exist in living cells as distinct foci by live-imaging fluorescently labeled CDK9, a kinase known to associate with active RNAPII. These foci were observed in different cell types derived from CDK9-mCherry knock-in mice. We show that these foci are very stable while highly dynamic in exchanging CDK9. Chromatin immunoprecipitation (ChIP) coupled with deep sequencing (ChIP-seq) data show that the genome-wide binding sites of CDK9 and initiating RNAPII overlap on transcribed genes. Immunostaining shows that CDK9-mCherry foci colocalize with RNAPII-Ser5P, much less with RNAPII-Ser2P, and not with CDK12 (a kinase reported to be involved in the Ser2 phosphorylation) or with splicing factor SC35. In conclusion, transcription factories exist in living cells, and initiation and elongation of transcripts takes place in different nuclear compartments.
Saturday, April 13, 2013
Coactivators enable glucocorticoid receptor recruitment to fine-tune estrogen receptor transcriptional responses
Coactivators enable glucocorticoid receptor recruitment to fine-tune estrogen receptor transcriptional responses:
Nuclear receptors (NRs) are central regulators of pathophysiological processes; however, how their responses intertwine is still not fully understood. The aim of this study was to determine whether and how steroid NRs can influence each other’s activity under co-agonist treatment. We used a unique system consisting of a multicopy integration of an estrogen receptor responsive unit that allows direct visualization and quantification of estrogen receptor alpha (ERα) DNA binding, co-regulator recruitment and transcriptional readout. We find that ERα DNA loading is required for other type I nuclear receptors to be co-recruited after dual agonist treatment. We focused on ERα/glucocorticoid receptor interplay and demonstrated that it requires steroid receptor coactivators (SRC-2, SRC-3) and the mediator component MED14. We then validated this cooperative interplay on endogenous target genes in breast cancer cells. Taken together, this work highlights another layer of mechanistic complexity through which NRs cross-talk with each other on chromatin under multiple hormonal stimuli.
Nuclear receptors (NRs) are central regulators of pathophysiological processes; however, how their responses intertwine is still not fully understood. The aim of this study was to determine whether and how steroid NRs can influence each other’s activity under co-agonist treatment. We used a unique system consisting of a multicopy integration of an estrogen receptor responsive unit that allows direct visualization and quantification of estrogen receptor alpha (ERα) DNA binding, co-regulator recruitment and transcriptional readout. We find that ERα DNA loading is required for other type I nuclear receptors to be co-recruited after dual agonist treatment. We focused on ERα/glucocorticoid receptor interplay and demonstrated that it requires steroid receptor coactivators (SRC-2, SRC-3) and the mediator component MED14. We then validated this cooperative interplay on endogenous target genes in breast cancer cells. Taken together, this work highlights another layer of mechanistic complexity through which NRs cross-talk with each other on chromatin under multiple hormonal stimuli.
Thursday, April 11, 2013
Balancing Selection on a Regulatory Region Exhibiting Ancient Variation That Predates Human–Neandertal Divergence
Balancing Selection on a Regulatory Region Exhibiting Ancient Variation That Predates Human–Neandertal Divergence:
by Omer Gokcumen, Qihui Zhu, Lubbertus C. F. Mulder, Rebecca C. Iskow, Christian Austermann, Christopher D. Scharer, Towfique Raj, Jeremy M. Boss, Shamil Sunyaev, Alkes Price, Barbara Stranger, Viviana Simon, Charles Lee
Ancient population structure shaping contemporary genetic variation has been recently appreciated and has important implications regarding our understanding of the structure of modern human genomes. We identified a ∼36-kb DNA segment in the human genome that displays an ancient substructure. The variation at this locus exists primarily as two highly divergent haplogroups. One of these haplogroups (the NE1 haplogroup) aligns with the Neandertal haplotype and contains a 4.6-kb deletion polymorphism in perfect linkage disequilibrium with 12 single nucleotide polymorphisms (SNPs) across diverse populations. The other haplogroup, which does not contain the 4.6-kb deletion, aligns with the chimpanzee haplotype and is likely ancestral. Africans have higher overall pairwise differences with the Neandertal haplotype than Eurasians do for this NE1 locus (p<10−15). Moreover, the nucleotide diversity at this locus is higher in Eurasians than in Africans. These results mimic signatures of recent Neandertal admixture contributing to this locus. However, an in-depth assessment of the variation in this region across multiple populations reveals that African NE1 haplotypes, albeit rare, harbor more sequence variation than NE1 haplotypes found in Europeans, indicating an ancient African origin of this haplogroup and refuting recent Neandertal admixture. Population genetic analyses of the SNPs within each of these haplogroups, along with genome-wide comparisons revealed significant FST (p = 0.00003) and positive Tajima's D (p = 0.00285) statistics, pointing to non-neutral evolution of this locus. The NE1 locus harbors no protein-coding genes, but contains transcribed sequences as well as sequences with putative regulatory function based on bioinformatic predictions and in vitro experiments. We postulate that the variation observed at this locus predates Human–Neandertal divergence and is evolving under balancing selection, especially among European populations.
by Omer Gokcumen, Qihui Zhu, Lubbertus C. F. Mulder, Rebecca C. Iskow, Christian Austermann, Christopher D. Scharer, Towfique Raj, Jeremy M. Boss, Shamil Sunyaev, Alkes Price, Barbara Stranger, Viviana Simon, Charles Lee
Ancient population structure shaping contemporary genetic variation has been recently appreciated and has important implications regarding our understanding of the structure of modern human genomes. We identified a ∼36-kb DNA segment in the human genome that displays an ancient substructure. The variation at this locus exists primarily as two highly divergent haplogroups. One of these haplogroups (the NE1 haplogroup) aligns with the Neandertal haplotype and contains a 4.6-kb deletion polymorphism in perfect linkage disequilibrium with 12 single nucleotide polymorphisms (SNPs) across diverse populations. The other haplogroup, which does not contain the 4.6-kb deletion, aligns with the chimpanzee haplotype and is likely ancestral. Africans have higher overall pairwise differences with the Neandertal haplotype than Eurasians do for this NE1 locus (p<10−15). Moreover, the nucleotide diversity at this locus is higher in Eurasians than in Africans. These results mimic signatures of recent Neandertal admixture contributing to this locus. However, an in-depth assessment of the variation in this region across multiple populations reveals that African NE1 haplotypes, albeit rare, harbor more sequence variation than NE1 haplotypes found in Europeans, indicating an ancient African origin of this haplogroup and refuting recent Neandertal admixture. Population genetic analyses of the SNPs within each of these haplogroups, along with genome-wide comparisons revealed significant FST (p = 0.00003) and positive Tajima's D (p = 0.00285) statistics, pointing to non-neutral evolution of this locus. The NE1 locus harbors no protein-coding genes, but contains transcribed sequences as well as sequences with putative regulatory function based on bioinformatic predictions and in vitro experiments. We postulate that the variation observed at this locus predates Human–Neandertal divergence and is evolving under balancing selection, especially among European populations.
Tuesday, April 9, 2013
Dual role for Hox genes and Hox co-factors in conferring leg motoneuron survival and identity in Drosophila [RESEARCH ARTICLES]
Dual role for Hox genes and Hox co-factors in conferring leg motoneuron survival and identity in Drosophila [RESEARCH ARTICLES]: Myungin Baek, Jonathan Enriquez, and Richard S. Mann
Adult Drosophila walk using six multi-jointed legs, each controlled by ~50 leg motoneurons (MNs). Although MNs have stereotyped morphologies, little is known about how they are specified. Here, we describe the function of Hox genes and homothorax (hth), which encodes a Hox co-factor, in Drosophila leg MN development. Removing either Hox or Hth function from a single neuroblast (NB) lineage results in MN apoptosis. A single Hox gene, Antennapedia (Antp), is primarily responsible for MN survival in all three thoracic segments. When cell death is blocked, partially penetrant axon branching errors are observed in Hox mutant MNs. When single MNs are mutant, errors in both dendritic and axon arborizations are observed. Our data also suggest that Antp levels in post-mitotic MNs are important for specifying their identities. Thus, in addition to being essential for survival, Hox and hth are required to specify accurate MN morphologies in a level-dependent manner.
Adult Drosophila walk using six multi-jointed legs, each controlled by ~50 leg motoneurons (MNs). Although MNs have stereotyped morphologies, little is known about how they are specified. Here, we describe the function of Hox genes and homothorax (hth), which encodes a Hox co-factor, in Drosophila leg MN development. Removing either Hox or Hth function from a single neuroblast (NB) lineage results in MN apoptosis. A single Hox gene, Antennapedia (Antp), is primarily responsible for MN survival in all three thoracic segments. When cell death is blocked, partially penetrant axon branching errors are observed in Hox mutant MNs. When single MNs are mutant, errors in both dendritic and axon arborizations are observed. Our data also suggest that Antp levels in post-mitotic MNs are important for specifying their identities. Thus, in addition to being essential for survival, Hox and hth are required to specify accurate MN morphologies in a level-dependent manner.
Thursday, April 4, 2013
[Report] Transposition-Driven Genomic Heterogeneity in the Drosophila Brain
[Report] Transposition-Driven Genomic Heterogeneity in the Drosophila Brain: Transposon movement in memory-relevant neurons in fruit flies increases neuronal diversity within and between animals.
Authors: Paola N. Perrat, Shamik DasGupta, Jie Wang, William Theurkauf, Zhiping Weng, Michael Rosbash, Scott Waddell
Authors: Paola N. Perrat, Shamik DasGupta, Jie Wang, William Theurkauf, Zhiping Weng, Michael Rosbash, Scott Waddell
[Report] Drosophila H1 Regulates the Genetic Activity of Heterochromatin by Recruitment of Su(var)3-9
[Report] Drosophila H1 Regulates the Genetic Activity of Heterochromatin by Recruitment of Su(var)3-9: The “fifth” histone, H1, acts to recruit a histone-methylating enzyme to silence specific regions of the genome.
Authors: Xingwu Lu, Sandeep N. Wontakal, Harsh Kavi, Byung Ju Kim, Paloma M. Guzzardo, Alexander V. Emelyanov, Na Xu, Gregory J. Hannon, Jiri Zavadil, Dmitry V. Fyodorov, Arthur I. Skoultchi
Authors: Xingwu Lu, Sandeep N. Wontakal, Harsh Kavi, Byung Ju Kim, Paloma M. Guzzardo, Alexander V. Emelyanov, Na Xu, Gregory J. Hannon, Jiri Zavadil, Dmitry V. Fyodorov, Arthur I. Skoultchi
Wednesday, April 3, 2013
Why I Study Duck Genitalia
Why I Study Duck Genitalia:
In the past few days, the Internet has been filled with commentary on whether the National Science Foundation should have paid for my study on duck genitalia, and 88.7 percent of respondents to a Fox news online poll agreed that studying duck genitalia is wasteful government spending. The commentary supporting and decrying the study continues to grow. As the lead investigator in this research, I would like to weigh in on the controversy and offer some insights into the process of research funding by the NSF.









In the past few days, the Internet has been filled with commentary on whether the National Science Foundation should have paid for my study on duck genitalia, and 88.7 percent of respondents to a Fox news online poll agreed that studying duck genitalia is wasteful government spending. The commentary supporting and decrying the study continues to grow. As the lead investigator in this research, I would like to weigh in on the controversy and offer some insights into the process of research funding by the NSF.






Tuesday, April 2, 2013
Promoter Sequence Determines the Relationship between Expression Level and Noise
Promoter Sequence Determines the Relationship between Expression Level and Noise:
by Lucas B. Carey, David van Dijk, Peter M. A. Sloot, Jaap A. Kaandorp, Eran Segal
The ability of cells to accurately control gene expression levels in response to extracellular cues is limited by the inherently stochastic nature of transcriptional regulation. A change in transcription factor (TF) activity results in changes in the expression of its targets, but the way in which cell-to-cell variability in expression (noise) changes as a function of TF activity, and whether targets of the same TF behave similarly, is not known. Here, we measure expression and noise as a function of TF activity for 16 native targets of the transcription factor Zap1 that are regulated by it through diverse mechanisms. For most activated and repressed Zap1 targets, noise decreases as expression increases. Kinetic modeling suggests that this is due to two distinct Zap1-mediated mechanisms that both change the frequency of transcriptional bursts. Notably, we found that another mechanism of repression by Zap1, which is encoded in the promoter DNA, likely decreases the size of transcriptional bursts, producing a unique transcriptional state characterized by low expression and low noise. In addition, we find that further reduction in noise is achieved when a single TF both activates and represses a single target gene. Our results suggest a global principle whereby at low TF concentrations, the dominant source of differences in expression between promoters stems from differences in burst frequency, whereas at high TF concentrations differences in burst size dominate. Taken together, we show that the precise amount by which noise changes with expression is specific to the regulatory mechanism of transcription and translation that acts at each gene.
by Lucas B. Carey, David van Dijk, Peter M. A. Sloot, Jaap A. Kaandorp, Eran Segal
The ability of cells to accurately control gene expression levels in response to extracellular cues is limited by the inherently stochastic nature of transcriptional regulation. A change in transcription factor (TF) activity results in changes in the expression of its targets, but the way in which cell-to-cell variability in expression (noise) changes as a function of TF activity, and whether targets of the same TF behave similarly, is not known. Here, we measure expression and noise as a function of TF activity for 16 native targets of the transcription factor Zap1 that are regulated by it through diverse mechanisms. For most activated and repressed Zap1 targets, noise decreases as expression increases. Kinetic modeling suggests that this is due to two distinct Zap1-mediated mechanisms that both change the frequency of transcriptional bursts. Notably, we found that another mechanism of repression by Zap1, which is encoded in the promoter DNA, likely decreases the size of transcriptional bursts, producing a unique transcriptional state characterized by low expression and low noise. In addition, we find that further reduction in noise is achieved when a single TF both activates and represses a single target gene. Our results suggest a global principle whereby at low TF concentrations, the dominant source of differences in expression between promoters stems from differences in burst frequency, whereas at high TF concentrations differences in burst size dominate. Taken together, we show that the precise amount by which noise changes with expression is specific to the regulatory mechanism of transcription and translation that acts at each gene.
Is junk DNA bunk? [Evolution]
Is junk DNA bunk? [Evolution]: Do data from the Encyclopedia Of DNA Elements (ENCODE) project render the notion of junk DNA obsolete? Here, I review older arguments for junk grounded in the C-value paradox and propose a thought experiment to challenge ENCODE’s ontology. Specifically, what would we expect for the number of functional elements (as...
The Border Between the Ultrabithorax and abdominal-A Regulatory Domains in the Drosophila Bithorax Complex [Gene Expression]
The Border Between the Ultrabithorax and abdominal-A Regulatory Domains in the Drosophila Bithorax Complex [Gene Expression]:
The bithorax complex in Drosophila melanogaster includes three homeobox-containing genes—Ultrabithorax (Ubx), abdominal-A (abd-A), and Abdominal-B (Abd-B)—which are required for the proper differentiation of the posterior 10 segments of the body. Each of these genes has multiple distinct regulatory regions; there is one for each segmental unit of the body plan where the genes are expressed. One additional protein- coding gene in the bithorax complex, Glut3, a sugar-transporter homolog, can be deleted without phenotype. We focus here on the upstream regulatory region for Ubx, the bithoraxoid (bxd) domain, and its border with the adjacent infraabdominal-2 (iab-2) domain, which controls abdA. These two domains can be defined by the phenotypes of rearrangement breakpoints, and by the expression patterns of enhancer traps. In D. virilis, the homeotic cluster is split between Ubx and abd-A, and so the border can also be located by a sequence comparison between species. When the border region is deleted in melanogaster, the flies show a dominant phenotype called Front-ultraabdominal (Fub); the first abdominal segment is transformed into a copy of the second abdominal segment. Thus, the border blocks the spread of activation from the bxd domain into the iab-2 domain.
The bithorax complex in Drosophila melanogaster includes three homeobox-containing genes—Ultrabithorax (Ubx), abdominal-A (abd-A), and Abdominal-B (Abd-B)—which are required for the proper differentiation of the posterior 10 segments of the body. Each of these genes has multiple distinct regulatory regions; there is one for each segmental unit of the body plan where the genes are expressed. One additional protein- coding gene in the bithorax complex, Glut3, a sugar-transporter homolog, can be deleted without phenotype. We focus here on the upstream regulatory region for Ubx, the bithoraxoid (bxd) domain, and its border with the adjacent infraabdominal-2 (iab-2) domain, which controls abdA. These two domains can be defined by the phenotypes of rearrangement breakpoints, and by the expression patterns of enhancer traps. In D. virilis, the homeotic cluster is split between Ubx and abd-A, and so the border can also be located by a sequence comparison between species. When the border region is deleted in melanogaster, the flies show a dominant phenotype called Front-ultraabdominal (Fub); the first abdominal segment is transformed into a copy of the second abdominal segment. Thus, the border blocks the spread of activation from the bxd domain into the iab-2 domain.
Monday, April 1, 2013
Highly conserved elements discovered in vertebrates are present in non-syntenic loci of tunicates, act as enhancers and can be transcribed during development
Highly conserved elements discovered in vertebrates are present in non-syntenic loci of tunicates, act as enhancers and can be transcribed during development:
Co-option of cis-regulatory modules has been suggested as a mechanism for the evolution of expression sites during development. However, the extent and mechanisms involved in mobilization of cis-regulatory modules remains elusive. To trace the history of non-coding elements, which may represent candidate ancestral cis-regulatory modules affirmed during chordate evolution, we have searched for conserved elements in tunicate and vertebrate (Olfactores) genomes. We identified, for the first time, 183 non-coding sequences that are highly conserved between the two groups. Our results show that all but one element are conserved in non-syntenic regions between vertebrate and tunicate genomes, while being syntenic among vertebrates. Nevertheless, in all the groups, they are significantly associated with transcription factors showing specific functions fundamental to animal development, such as multicellular organism development and sequence-specific DNA binding. The majority of these regions map onto ultraconserved elements and we demonstrate that they can act as functional enhancers within the organism of origin, as well as in cross-transgenesis experiments, and that they are transcribed in extant species of Olfactores. We refer to the elements as ‘Olfactores conserved non-coding elements’.
Co-option of cis-regulatory modules has been suggested as a mechanism for the evolution of expression sites during development. However, the extent and mechanisms involved in mobilization of cis-regulatory modules remains elusive. To trace the history of non-coding elements, which may represent candidate ancestral cis-regulatory modules affirmed during chordate evolution, we have searched for conserved elements in tunicate and vertebrate (Olfactores) genomes. We identified, for the first time, 183 non-coding sequences that are highly conserved between the two groups. Our results show that all but one element are conserved in non-syntenic regions between vertebrate and tunicate genomes, while being syntenic among vertebrates. Nevertheless, in all the groups, they are significantly associated with transcription factors showing specific functions fundamental to animal development, such as multicellular organism development and sequence-specific DNA binding. The majority of these regions map onto ultraconserved elements and we demonstrate that they can act as functional enhancers within the organism of origin, as well as in cross-transgenesis experiments, and that they are transcribed in extant species of Olfactores. We refer to the elements as ‘Olfactores conserved non-coding elements’.
Subscribe to:
Posts (Atom)