Friday, December 31, 2010
Booze myths: The truth at the bottom of the bottle
Thursday, December 30, 2010
Nucleosome-mediated cooperativity between transcription factors [Biophysics_And_Computational_Biology]
Cooperative binding of transcription factors (TFs) to promoters and other regulatory regions is essential for precise gene expression. The classical model of cooperativity requires direct interactions between TFs, thus constraining the arrangement of TF sites in regulatory regions. Recent genomic and functional studies, however, demonstrate a great deal of flexibility in such arrangements with variable distances, numbers of sites, and identities of TF sites located in cis-regulatory regions. Such flexibility is inconsistent with cooperativity by direct interactions between TFs. Here, we demonstrate that strong cooperativity among noninteracting TFs can be achieved by their competition with nucleosomes. We find that the mechanism of nucleosome-mediated cooperativity is analogous to cooperativity in another multimolecular complex: hemoglobin. This surprising analogy provides deep insights, with parallels between the heterotropic regulation of hemoglobin (e.g., the Bohr effect) and the roles of nucleosome-positioning sequences and chromatin modifications in gene expression. Nucleosome-mediated cooperativity is consistent with several experimental studies, is equally applicable to repressors and activators, allows substantial flexibility in and modularity of regulatory regions, and provides a rationale for a broad range of genomic and evolutionary observations. Striking parallels between cooperativity in hemoglobin and in transcriptional regulation point to a general mechanism that can be used in various biological systems.
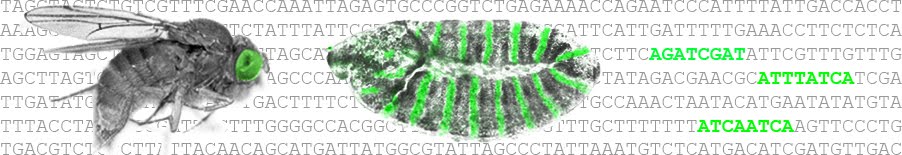
"[Research Article] Identification of Functional Elements and Regulatory Circuits by Drosophila modENCODE
Authors: The modENCODE Consortium, Sushmita Roy, Jason Ernst, Peter V. Kharchenko, Pouya Kheradpour, Nicolas Negre, Matthew L. Eaton, Jane M. Landolin, Christopher A. Bristow, Lijia Ma, Michael F. Lin, Stefan Washietl, Bradley I. Arshinoff, Ferhat Ay, Patrick E. Meyer, Nicolas Robine, Nicole L. Washington, Luisa Di Stefano, Eugene Berezikov, Christopher D. Brown, Rogerio Candeias, Joseph W. Carlson, Adrian Carr, Irwin Jungreis, Daniel Marbach, Rachel Sealfon, Michael Y. Tolstorukov, Sebastian Will, Artyom A. Alekseyenko, Carlo Artieri, Benjamin W. Booth, Angela N. Brooks, Qi Dai, Carrie A. Davis, Michael O. Duff, Xin Feng, Andrey A. Gorchakov, Tingting Gu, Jorja G. Henikoff, Philipp Kapranov, Renhua Li, Heather K. MacAlpine, John Malone, Aki Minoda, Jared Nordman, Katsutomo Okamura, Marc Perry, Sara K. Powell, Nicole C. Riddle, Akiko Sakai, Anastasia Samsonova, Jeremy E. Sandler, Yuri B. Schwartz, Noa Sher, Rebecca Spokony, David Sturgill, Marijke van Baren, Kenneth H. Wan, Li Yang, Charles Yu, Elise Feingold, Peter Good, Mark Guyer, Rebecca Lowdon, Kami Ahmad, Justen Andrews, Bonnie Berger, Steven E. Brenner, Michael R. Brent, Lucy Cherbas, Sarah C. R. Elgin, Thomas R. Gingeras, Robert Grossman, Roger A. Hoskins, Thomas C. Kaufman, William Kent, Mitzi I. Kuroda, Terry Orr-Weaver, Norbert Perrimon, Vincenzo Pirrotta, James W. Posakony, Bing Ren, Steven Russell, Peter Cherbas, Brenton R. Graveley, Suzanna Lewis, Gos Micklem, Brian Oliver, Peter J. Park, Susan E. Celniker, Steven Henikoff, Gary H. Karpen, Eric C. Lai, David M. MacAlpine, Lincoln D. Stein, Kevin P. White, Manolis Kellis"
[Research Article] Integrative Analysis of the Caenorhabditis elegans Genome by the modENCODE Project
Authors: Mark B. Gerstein, Zhi John Lu, Eric L. Van Nostrand, Chao Cheng, Bradley I. Arshinoff, Tao Liu, Kevin Y. Yip, Rebecca Robilotto, Andreas Rechtsteiner, Kohta Ikegami, Pedro Alves, Aurelien Chateigner, Marc Perry, Mitzi Morris, Raymond K. Auerbach, Xin Feng, Jing Leng, Anne Vielle, Wei Niu, Kahn Rhrissorrakrai, Ashish Agarwal, Roger P. Alexander, Galt Barber, Cathleen M. Brdlik, Jennifer Brennan, Jeremy Jean Brouillet, Adrian Carr, Ming-Sin Cheung, Hiram Clawson, Sergio Contrino, Luke O. Dannenberg, Abby F. Dernburg, Arshad Desai, Lindsay Dick, Andréa C. Dosé, Jiang Du, Thea Egelhofer, Sevinc Ercan, Ghia Euskirchen, Brent Ewing, Elise A. Feingold, Reto Gassmann, Peter J. Good, Phil Green, Francois Gullier, Michelle Gutwein, Mark S. Guyer, Lukas Habegger, Ting Han, Jorja G. Henikoff, Stefan R. Henz, Angie Hinrichs, Heather Holster, Tony Hyman, A. Leo Iniguez, Judith Janette, Morten Jensen, Masaomi Kato, W. James Kent, Ellen Kephart, Vishal Khivansara, Ekta Khurana, John K. Kim, Paulina Kolasinska-Zwierz, Eric C. Lai, Isabel Latorre, Amber Leahey, Suzanna Lewis, Paul Lloyd, Lucas Lochovsky, Rebecca F. Lowdon, Yaniv Lubling, Rachel Lyne, Michael MacCoss, Sebastian D. Mackowiak, Marco Mangone, Sheldon McKay, Desirea Mecenas, Gennifer Merrihew, David M. Miller III, Andrew Muroyama, John I. Murray, Siew-Loon Ooi, Hoang Pham, Taryn Phippen, Elicia A. Preston, Nikolaus Rajewsky, Gunnar Rätsch, Heidi Rosenbaum, Joel Rozowsky, Kim Rutherford, Peter Ruzanov, Mihail Sarov, Rajkumar Sasidharan, Andrea Sboner, Paul Scheid, Eran Segal, Hyunjin Shin, Chong Shou, Frank J. Slack, Cindie Slightam, Richard Smith, William C. Spencer, E. O. Stinson, Scott Taing, Teruaki Takasaki, Dionne Vafeados, Ksenia Voronina, Guilin Wang, Nicole L. Washington, Christina M. Whittle, Beijing Wu, Koon-Kiu Yan, Georg Zeller, Zheng Zha, Mei Zhong, Xingliang Zhou, modENCODE Consortium, Julie Ahringer, Susan Strome, Kristin C. Gunsalus, Gos Micklem, X. Shirley Liu, Valerie Reinke, Stuart K. Kim, LaDeana W. Hillier, Steven Henikoff, Fabio Piano, Michael Snyder, Lincoln Stein, Jason D. Lieb, Robert H. Waterston"
FOXA1 is a key determinant of estrogen receptor function and endocrine response
FOXA1 is a key determinant of estrogen receptor function and endocrine response
Nature Genetics 43, 27 (2011). doi:10.1038/ng.730
Authors: Antoni Hurtado, Kelly A Holmes, Caryn S Ross-Innes, Dominic Schmidt & Jason S Carroll
Transcriptional activation of polycomb-repressed genes by ZRF1
Transcriptional activation of polycomb-repressed genes by ZRF1
Nature 468, 7327 (2010). doi:10.1038/nature09574
Authors: Holger Richly, Luciana Rocha-Viegas, Joana Domingues Ribeiro, Santiago Demajo, Gunes Gundem, Nuria Lopez-Bigas, Tekeya Nakagawa, Sabine Rospert, Takashi Ito & Luciano Di Croce
Covalent modification of histones is fundamental in orchestrating chromatin dynamics and transcription. One example of such an epigenetic mark is the mono-ubiquitination of histones, which mainly occurs at histone H2A and H2B. Ubiquitination of histone H2A has been implicated in polycomb-mediated transcriptional silencing. However, the precise role of the ubiquitin mark during silencing is still elusive. Here we show in human cell lines that ZRF1 (zuotin-related factor 1) is specifically recruited to histone H2A when it is ubiquitinated at Lys 119 by means of a novel ubiquitin-interacting domain that is located in the evolutionarily conserved zuotin domain. At the onset of differentiation, ZRF1 specifically displaces polycomb-repressive complex 1 (PRC1) from chromatin and facilitates transcriptional activation. A genome-wide mapping of ZRF1, RING1B and H2A-ubiquitin targets revealed its involvement in the regulation of a large set of polycomb target genes, emphasizing the key role ZRF1 has in cell fate decisions. We provide here a model of the molecular mechanism of switching polycomb-repressed genes to an active state.
Phenotypic Landscape of a Bacterial Cell
Wednesday, December 29, 2010
Novel genes exhibit distinct patterns of function acquisition and network integration
Genes are created by a variety of evolutionary processes, some of which generate duplicate copies of an entire gene, while others rearrange pre-existing genetic elements or co-opt previously non-coding sequence to create genes with 'novel' sequences. These novel genes are thought to contribute to distinct phenotypes that distinguish organisms. The creation, evolution, and function of duplicated genes are well-studied; however, the genesis and early evolution of novel genes are not well-characterized. We developed a computational approach to investigate these issues by integrating genome-wide comparative phylogenetic analysis with functional and interaction data derived from small-scale and high-throughput experiments.
Results:
We examine the function and evolution of new genes in the yeast Saccharomyces cerevisiae. We observed significant differences in the functional attributes and interactions of genes created at different times and by different mechanisms. Novel genes are initially less integrated into cellular networks than duplicate genes, but they appear to gain functions and interactions more quickly than duplicates. Recently created duplicated genes show evidence of adapting existing functions to environmental changes, while young novel genes do not exhibit enrichment for any particular functions. Finally, we found a significant preference for genes to interact with other genes of similar age and origin.
Conclusions:
Our results suggest a strong relationship between how and when genes are created and the roles they play in the cell. Overall, genes tend to become more integrated into the functional networks of the cell with time, but the dynamics of this process differ significantly between duplicate and novel genes."
Transcriptional Control of Photosynthesis Genes: The Evolutionarily Conserved Regulatory Mechanism in Plastid Genome Function
Chloroplast sensor kinase (CSK) is a bacterial-type sensor histidine kinase found in chloroplasts—photosynthetic plastids—in eukaryotic plants and algae. Using a yeast two-hybrid screen, we demonstrate recognition and interactions between: CSK, plastid transcription kinase (PTK), and a bacterial-type RNA polymerase sigma factor-1 (SIG-1). CSK interacts with itself, with SIG-1, and with PTK. PTK also interacts directly with SIG-1. PTK has previously been shown to catalyze phosphorylation of plastid-encoded RNA polymerase (PEP), suppressing plastid transcription nonspecifically. Phospho-PTK is inactive as a PEP kinase. Here, we propose that phospho-CSK acts as a PTK kinase, releasing PTK repression of chloroplast transcription, while CSK also acts as a SIG-1 kinase, blocking transcription specifically at the gene promoter of chloroplast photosystem I. Oxidation of the photosynthetic electron carrier plastoquinone triggers phosphorylation of CSK, inducing chloroplast photosystem II while suppressing photosystem I. CSK places photosystem gene transcription under the control of photosynthetic electron transport. This redox signaling pathway has its origin in cyanobacteria, photosynthetic prokaryotes from which chloroplasts evolved. The persistence of this mechanism in cytoplasmic organelles of photosynthetic eukaryotes is in precise agreement with the CoRR hypothesis for the function of organellar genomes: the plastid genome and its primary gene products are Co-located for Redox Regulation. Genes are retained in plastids primarily in order for their expression to be subject to this rapid and robust redox regulatory transcriptional control mechanism, whereas plastid genes also encode genetic system components, such as some ribosomal proteins and RNAs, that exist in order to support this primary, redox regulatory control of photosynthesis genes. Plastid genome function permits adaptation of the photosynthetic apparatus to changing environmental conditions of light quantity and quality.
"Ancestral Regulatory Circuits Governing Ectoderm Patterning Downstream of Nodal and BMP2/4 Revealed by Gene Regulatory Network Analysis in an Echinoderm
Echinoderms (sea urchins, starfish, etc.) are marine invertebrates that share a close ancestry with vertebrates. Their embryos offer many advantages for the analysis of transcriptional circuits that control developmental programs. During early development of the common sea urchin Paracentrotus lividus, a signaling center located within the ventral ectoderm sends two key signals, Nodal and BMP2/4, that control patterning of the embryo along the whole dorsal-ventral axis. How this signaling center works is not understood. We have conducted a large-scale functional analysis of the genes responsible for patterning of the ectoderm along the dorsal-ventral axis. We identified direct targets of Nodal and BMP2/4 and identified several key regulators that mediate the effects of these factors and drive essential and probably ancient regulatory circuits that together constitute a transcriptional program controlling morphogenesis of the embryo. In addition, we uncovered a striking parallel between the mouse embryo and the sea urchin embryo by showing that in both models a neurogenic ectoderm is the default state of ectoderm differentiation in the absence of Nodal and BMP signaling. Our results support the idea that inhibition of Nodal and BMP signaling was probably an ancient mechanism to specify neural cells in the ancestor of vertebrates.
Initiator Elements Function to Determine the Activity State of BX-C Enhancers
Understanding how genes become activated is one of the primary areas of research in modern biology. In order to decipher the DNA components required for this process, scientists have traditionally turned to transgenic reporter assays, where DNA elements are removed from their native environment and placed next to a simplified reporter gene to monitor transcriptional activation. Although this approach is powerful, it can result in artifacts stemming from the channelization of regulatory element activities into predetermined classes. In this manuscript, we investigate the biological role of elements from the Drosophila bithorax complex, called initiators. In transgenic assays, these elements have been categorized as enhancers. However, genetic analysis suggests that, in situ, these elements perform a far more complex function. Here, using a new method to repeatedly target a genetic locus for mutagenesis, we show that initiators function as control elements that coordinate the activity of nearby enhancers and silencers. Overall, our study highlights how gene expression can be controlled through a hierarchical arrangement of cis-regulatory elements.
A Young Drosophila Duplicate Gene Plays Essential Roles in Spermatogenesis by Regulating Several Y-Linked Male Fertility Genes
Gene duplication has long been appreciated as a major source for new genes and new functions. Nevertheless, it is still a fascinating mystery how new duplicate genes are functionally integrated into the existing gene network and how they contribute to the novel functions of organisms at the pathway level. By studying the recently originated kep1 gene family in Drosophila melanogaster, we show that one of the young duplicate genes, nsr, has evolved important biological functions associated with male reproduction by regulating several essential fertility genes in the short evolutionary period after its birth. The evolutionary dynamics, biological roles, and the underlying molecular mechanism of nsr revealed in this study present a vivid and comprehensive example of how new genes acquire important biological functions and demonstrate that recently originated new genes can regulate pre-existing essential genes and create novel architectures of genetic pathways.
Tuesday, December 21, 2010
Bimodal gene expression in noncooperative regulatory systems [Applied_Mathematics]
Bimodality of gene expression, as a mechanism contributing to phenotypic diversity, enhances the survival of cells in a fluctuating environment. To date, the bimodal response of a gene regulatory system has been attributed to the cooperativity of transcription factor binding or to feedback loops. It has remained unclear whether noncooperative binding of transcription factors can give rise to bimodality in an open-loop system. We study a theoretical model of gene expression in a two-step cascade (a deterministically monostable system) in which the regulatory gene produces transcription factors that have a nonlinear effect on the activity of the target gene. We show that a unimodal distribution of transcription factors over the cell population can generate a bimodal steady-state output without cooperative transcription factor binding. We introduce a simple method of geometric construction that allows one to predict the onset of bimodality. The construction only involves the parameters of bursting of the regulatory gene and the dose–response curve of the target gene. Using this method, we show that the gene expression may switch between unimodal and bimodal as the concentration of inducers or corepressors is varied. These findings may explain the experimentally observed bimodal response of cascades consisting of a fluorescent protein reporter controlled by the tetracycline repressor. The geometric construction provides a useful tool for designing experiments and for interpretation of their results. Our findings may have important implications for understanding the strategies adopted by cell populations to survive in changing environments.
"Friday, December 17, 2010
A Single Enhancer Regulating the Differential Expression of Duplicated Red-Sensitive Opsin Genes in Zebrafish
Among vertebrates, fish may have the most advanced color vision. They have greatly varied repertoires of color sensors called visual opsins, possibly reflecting evolutionary adaptation to their diverse photic environments in water, and are an excellent model to study the evolution of vertebrate color vision. This is achieved by multiplying opsin genes and differentiating their absorption light spectra and expression patterns. However, little is understood regarding how the opsin genes are regulated to achieve the differential expression pattern. In this study, we focused on the duplicated red-sensitive opsin genes of zebrafish to tackle this problem. We discovered an “enhancer” region near the two red opsin genes that plays a crucial role in their differential expression pattern. Our results suggest that the two red opsin genes interact with the enhancer competitively in a developmentally restricted manner. Sharing a regulatory region could be a general way to facilitate the expression differentiation in duplicated visual opsin genes.
[Report] New Genes in Drosophila Quickly Become Essential
Authors: Sidi Chen, Yong E. Zhang, Manyuan Long"
Thursday, December 16, 2010
Natural Variation, Functional Pleiotropy and Transcriptional Contexts of Odorant Binding Protein Genes in Drosophila melanogaster [Genome and systems biology]
How functional diversification affects the organization of the transcriptome is a central question in systems genetics. To explore this issue, we sequenced all six Odorant binding protein (Obp) genes located on the X chromosome, four of which occur as a cluster, in 219 inbred wild-derived lines of Drosophila melanogaster and tested for associations between genetic and phenotypic variation at the organismal and transcriptional level. We observed polymorphisms in Obp8a, Obp19a, Obp19b, and Obp19c associated with variation in olfactory responses and polymorphisms in Obp19d associated with variation in life span. We inferred the transcriptional context, or 'niche,' of each gene by identifying expression polymorphisms where genetic variation in these Obp genes was associated with variation in expression of transcripts genetically correlated to each Obp gene. All six Obp genes occupied a distinct transcriptional niche. Gene ontology enrichment analysis revealed associations of different Obp transcriptional niches with olfactory behavior, synaptic transmission, detection of signals regulating tissue development and apoptosis, postmating behavior and oviposition, and nutrient sensing. Our results show that diversification of the Obp family has organized distinct transcriptional niches that reflect their acquisition of additional functions.
"Evolution of the Drosophila Feminizing Switch Gene Sex-lethal [Developmental and behavioral genetics]
In Drosophila melanogaster, the gene Sex-lethal (Sxl) controls all aspects of female development. Since melanogaster males lacking Sxl appear wild type, Sxl would seem to be functionally female specific. Nevertheless, in insects as diverse as honeybees and houseflies, Sxl seems not to determine sex or to be functionally female specific. Here we describe three lines of work that address the questions of how, when, and even whether the ancestor of melanogaster Sxl ever shed its non-female-specific functions. First, to test the hypothesis that the birth of Sxl's closest paralog allowed Sxl to lose essential ancestral non-female-specific functions, we determined the CG3056 null phenotype. That phenotype failed to support this hypothesis. Second, to define when Sxl might have lost ancestral non-female-specific functions, we isolated and characterized Sxl mutations in D. virilis, a species distant from melanogaster and notable for the large amount of Sxl protein expression in males. We found no change in Sxl regulation or functioning in the 40+ MY since these two species diverged. Finally, we discovered conserved non-sex-specific Sxl mRNAs containing a previously unknown, potentially translation-initiating exon, and we identified a conserved open reading frame starting in Sxl male-specific exon 3. We conclude that Drosophila Sxl may appear functionally female specific not because it lost non-female-specific functions, but because those functions are nonessential in the laboratory. The potential evolutionary relevance of these nonessential functions is discussed.
"Intercalation of a new tier of transcription regulation into an ancient circuit
Intercalation of a new tier of transcription regulation into an ancient circuit
Nature 468, 7326 (2010). doi:10.1038/nature09560
Authors: Lauren N. Booth, Brian B. Tuch & Alexander D. Johnson
Changes in gene regulatory networks are a major source of evolutionary novelty. Here we describe a specific type of network rewiring event, one that intercalates a new level of transcriptional control into an ancient circuit. We deduce that, over evolutionary time, the direct ancestral connections between a regulator and its target genes were broken and replaced by indirect connections, preserving the overall logic of the ancestral circuit but producing a new behaviour. The example was uncovered through a series of experiments in three ascomycete yeasts: the bakers’ yeast Saccharomyces cerevisiae, the dairy yeast Kluyveromyces lactis and the human pathogen Candida albicans. All three species have three cell types: two mating-competent cell forms (a and α) and the product of their mating (a/α), which is mating-incompetent. In the ancestral mating circuit, two homeodomain proteins, Mata1 and Matα2, form a heterodimer that directly represses four genes that are expressed only in a and α cells and are required for mating. In a relatively recent ancestor of K. lactis, a reorganization occurred. The Mata1–Matα2 heterodimer represses the same four genes (known as the core haploid-specific genes) but now does so indirectly through an intermediate regulatory protein, Rme1. The overall logic of the ancestral circuit is preserved (haploid-specific genes ON in a and α cells and OFF in a/α cells), but a new phenotype was produced by the rewiring: unlike S. cerevisiae and C. albicans, K. lactis integrates nutritional signals, by means of Rme1, into the decision of whether or not to mate.
Regulatory polymorphisms in the bovine Ankyrin 1 gene promoter are associated with tenderness and intra-muscular fat content
Recent QTL and gene expression studies have highlighted ankyrins as positional and functional candidate genes for meat quality. Our objective was to characterise the promoter region of the bovine ankyrin 1 gene and to test polymorphisms for association with sensory and technological meat quality measures.
Results:
Seven novel promoter SNPs were identified in a 1.11 kb region of the ankyrin 1 promoter in Angus, Charolais and Limousin bulls (n=15 per breed) as well as 141 crossbred beef animals for which meat quality data was available. Eighteen haplotypes were inferred with significant breed variation in haplotype frequencies. The five most frequent SNPs and the four most frequent haplotypes were subsequently tested for association with sensory and technological measures of meat quality in the crossbred population. SNP1, SNP3 and SNP4 (which were subsequently designated regulatory SNPs) and SNP5 were associated with traits that contribute to sensorial and technological measurements of tenderness and texture; Haplotype 1 and haplotype 4 were oppositely correlated with traits contributing to tenderness (P<0.05 for all). While no single SNP was associated with intramuscular fat (IMF), a clear association with increased IMF and juiciness was observed for haplotype 2.
Conclusion:
The conclusion from this study is that alleles defining haplotypes 2 and 4 could usefully contribute to marker SNP panels used to select individuals with improved IMF/juiciness or tenderness in a genome-assisted selection framework."
Wednesday, December 15, 2010
Evolution of an antifreeze protein by neofunctionalization under escape from adaptive conflict [Evolution]
The evolutionary model escape from adaptive conflict (EAC) posits that adaptive conflict between the old and an emerging new function within a single gene could drive the fixation of gene duplication, where each duplicate can freely optimize one of the functions. Although EAC has been suggested as a common process in functional evolution, definitive cases of neofunctionalization under EAC are lacking, and the molecular mechanisms leading to functional innovation are not well-understood. We report here clear experimental evidence for EAC-driven evolution of type III antifreeze protein gene from an old sialic acid synthase (SAS) gene in an Antarctic zoarcid fish. We found that an SAS gene, having both sialic acid synthase and rudimentary ice-binding activities, became duplicated. In one duplicate, the N-terminal SAS domain was deleted and replaced with a nascent signal peptide, removing pleiotropic structural conflict between SAS and ice-binding functions and allowing rapid optimization of the C-terminal domain to become a secreted protein capable of noncolligative freezing-point depression. This study reveals how minor functionalities in an old gene can be transformed into a distinct survival protein and provides insights into how gene duplicates facing presumed identical selection and mutation pressures at birth could take divergent evolutionary paths.
"Tuesday, December 14, 2010
Sleepy bees slur their waggle dance moves
If you extrapolate these results to humans, I now have something to blame it on!!!
Human Transcriptional Interactome of Chromatin Contribute to Gene Co-expression
Transcriptional interactome of chromatin is one of the important mechanisms in gene transcription regulation. By chromatin conformation capture and 3D FISH experiments, several chromatin interactions cases among sequence-distant genes or even inter-chromatin genes were reported. However, on genomics level, there is still little evidence to support these mechanisms. Recently based on Hi-C experiment, a genome-wide picture of chromatin interactions in human cells was presented. It provides a useful material for analysing whether the mechanism of transcriptional interactome is common.
Results:
The main work here is to demonstrate whether the effects of transcriptional interactome on gene co-expression exist on genomic level. While controlling the effects of transcription factors control similarities (TCS), we tested the correlation between Hi-C interaction and the mutual ranks of gene co-expression rates (provided by COXPRESdb) of intra-chromatin gene pairs. We used 6,084 genes with both TF annotation and co-expression information, and matched them into 273,458 pairs with similar Hi-C interaction ranks in different cell types. The results illustrate that co-expression is strongly associated with chromatin interaction. Further analysis using GO annotation reveals potential correlation between gene function similarity, Hi-C interaction and their co-expression.
Conclusions:
According to the results in this research, the intra-chromatin interactome may have relation to gene function and associate with co-expression. This study provides evidence for illustrating the effect of transcriptional interactome on transcription regulation."
Monday, December 13, 2010
Old and Wise: Why Do Smarter People Live Longer?
Intelligent people live longer--the correlation is as strong as that between smoking and premature death. But the reason is not fully understood. Beyond simply making wiser choices in life, these people also may have biology working in their favor. Now research in honeybees offers evidence that learning ability is indeed linked with a general capacity to withstand one of the rigors of aging--namely, oxidative stress. [More]"
The juries still out on which side of the line I fall...
2R and remodeling of vertebrate signal transduction engine
Whole genome duplication (WGD) is a special case of gene duplication, observed rarely in animals, where all genes duplicate simultaneously through polyploidisation. Two rounds of WGD (2R-WGD) occurred at the base of vertebrates, giving rise to an enormous wave of genetic novelty, but a systematic analysis of functional consequences of this event has not yet been performed. Results: We show that 2R-WGD affected overwhelming majority (74%) of signaling genes, in particular developmental pathways involving receptor tyrosine kinases, Wnt and TGF-beta ligands, GPCRs, and the apoptosis pathway. 2R-retained genes, in contrast to tandem duplicates, were enriched in protein interaction domains, and multifunctional signaling modules of Ras and MAP-kinase cascades. 2R-WGD had a fundamental impact on the cell-cycle machinery; redefined molecular building blocks of the neuronal synapse; and was formative for vertebrate brains. We investigated 2R-associated nodes in context of the human signaling network, as well as an inferred ancestral pre-2R (AP2R) network, and found that hubs (particularly involving negative regulations), were preferentially retained, with high-connectivity driving retention. Finally, microarrays and proteomics demonstrated a trend for gradual paralog expression divergence, independent of the duplication mechanism; but inferred ancestral expression states suggested preferential sub-functionalisation among 2R-ohnologs (2ROs). Conclusions: The 2R event left an indelible imprint on vertebrate signaling and cell-cycle. We show that 2R-WGD preferentially retained genes are associated with higher organismal complexity (e.g. locomotion, nervous system, morphogenesis), while genes associated with basic cellular functions (e.g. translation, replication, splicing, recombination; with the notable exception of cell-cycle) tended to be excluded. 2R-WGD set the stage for the emergence of key vertebrate functional novelties (such as complex brains, circulatory system, heart, bone, cartilage, musculature, and the adipose tissue). Full explanation of the impact of 2R on evolution, function, and the flow of information in vertebrate signaling networks is likely to have practical consequences for regenerative medicine, stem cell therapies, and cancer treatment."
How the vertebrates were made: selective pruning of a double-duplicated genome
Friday, December 10, 2010
Functional Analysis of CTCF During Mammalian Limb Development
Thursday, December 9, 2010
A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns
A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns
Nature 468, 7325 (2010). doi:10.1038/nature09632
Authors: Tomislav Domazet-Lošo & Diethard Tautz
Parallels between phylogeny and ontogeny have been discussed for almost two centuries, and a number of theories have been proposed to explain such patterns. Especially elusive is the phylotypic stage, a phase during development where species within a phylum are particularly similar to each other. Although this has formerly been interpreted as a recapitulation of phylogeny, it is now thought to reflect an ontogenetic progression phase, where strong constraints on developmental regulation and gene interactions exist. Several studies have shown that genes expressed during this stage evolve at a slower rate, but it has so far not been possible to derive an unequivocal molecular signature associated with this stage. Here we use a combination of phylostratigraphy and stage-specific gene expression data to generate a cumulative index that reflects the evolutionary age of the transcriptome at given ontogenetic stages. Using zebrafish ontogeny and adult development as a model, we find that the phylotypic stage does indeed express the oldest transcriptome set and that younger sets are expressed during early and late development, thus faithfully mirroring the hourglass model of morphological divergence. Reproductively active animals show the youngest transcriptome, with major differences between males and females. Notably, ageing animals express increasingly older genes. Comparisons with similar data sets from flies and nematodes show that this pattern occurs across phyla. Our results indicate that an old transcriptome marks the phylotypic phase and that phylogenetic differences at other ontogenetic stages correlate with the expression of newly evolved genes.
Gene expression divergence recapitulates the developmental hourglass model
Gene expression divergence recapitulates the developmental hourglass model
Nature 468, 7325 (2010). doi:10.1038/nature09634
Authors: Alex T. Kalinka, Karolina M. Varga, Dave T. Gerrard, Stephan Preibisch, David L. Corcoran, Julia Jarrells, Uwe Ohler, Casey M. Bergman & Pavel Tomancak
The observation that animal morphology tends to be conserved during the embryonic phylotypic period (a period of maximal similarity between the species within each animal phylum) led to the proposition that embryogenesis diverges more extensively early and late than in the middle, known as the hourglass model. This pattern of conservation is thought to reflect a major constraint on the evolution of animal body plans. Despite a wealth of morphological data confirming that there is often remarkable divergence in the early and late embryos of species from the same phylum, it is not yet known to what extent gene expression evolution, which has a central role in the elaboration of different animal forms, underpins the morphological hourglass pattern. Here we address this question using species-specific microarrays designed from six sequenced Drosophila species separated by up to 40 million years. We quantify divergence at different times during embryogenesis, and show that expression is maximally conserved during the arthropod phylotypic period. By fitting different evolutionary models to each gene, we show that at each time point more than 80% of genes fit best to models incorporating stabilizing selection, and that for genes whose evolutionarily optimal expression level is the same across all species, selective constraint is maximized during the phylotypic period. The genes that conform most to the hourglass pattern are involved in key developmental processes. These results indicate that natural selection acts to conserve patterns of gene expression during mid-embryogenesis, and provide a genome-wide insight into the molecular basis of the hourglass pattern of developmental evolution.
Wednesday, December 8, 2010
Rapid evolution of cold tolerance in stickleback
Climate change is predicted to lead to increased average temperatures and greater intensity and frequency of high and low temperature extremes, but the evolutionary consequences for biological communities are not well understood. Studies of adaptive evolution of temperature tolerance have typically involved correlative analyses of natural populations or artificial selection experiments in the laboratory. Field experiments are required to provide estimates of the timing and strength of natural selection, enhance understanding of the genetics of adaptation and yield insights into the mechanisms driving evolutionary change. Here, we report the experimental evolution of cold tolerance in natural populations of threespine stickleback fish (Gasterosteus aculeatus). We show that freshwater sticklebacks are able to tolerate lower minimum temperatures than marine sticklebacks and that this difference is heritable. We transplanted marine sticklebacks to freshwater ponds and measured the rate of evolution after three generations in this environment. Cold tolerance evolved at a rate of 0.63 haldanes to a value 2.5°C lower than that of the ancestral population, matching values found in wild freshwater populations. Our results suggest that cold tolerance is under strong selection and that marine sticklebacks carry sufficient genetic variation to adapt to changes in temperature over remarkably short time scales.
"Tuesday, December 7, 2010
Integration of RNA processing and expression level control modulates the function of the Drosophila Hox gene Ultrabithorax during adult development [RESEARCH ARTICLE]
Although most metazoan genes undergo alternative splicing, the functional relevance of the majority of alternative splicing products is still unknown. Here we explore this problem in the Drosophila Hox gene Ultrabithorax (Ubx). Ubx produces a family of six protein isoforms through alternative splicing. To investigate the functional specificity of the Ubx isoforms, we studied their role during the formation of the Drosophila halteres, small dorsal appendages that are essential for normal flight. Our work shows that isoform Ia, which is encoded by all Ubx exons, is more efficient than isoform IVa, which lacks the amino acids coded by two small exons, in controlling haltere development and regulating Ubx downstream targets. However, our experiments also demonstrate that the functional differences among the Ubx isoforms can be compensated for by increasing the expression levels of the less efficient form. The analysis of the DNA-binding profiles of Ubx isoforms to a natural Ubx target, spalt, shows no major differences in isoform DNA-binding activities, suggesting that alternative splicing might primarily affect the regulatory capacity of the isoforms rather than their DNA-binding patterns. Our results suggest that to obtain distinct functional outputs during normal development genes must integrate the generation of qualitative differences by alternative splicing to quantitative processes affecting isoform protein expression levels.
"Monday, December 6, 2010
Assessing Computational Methods of Cis-Regulatory Module Prediction
Transcriptional regulation involves multiple transcription factors binding to DNA sequences. A limited repertoire of transcription factors performs this complex regulatory step through various spatial and temporal interactions between themselves and their binding sites. These transcription factor binding interactions are clustered as distinct modules: cis-regulatory modules (CRMs). Computational methods attempting to identify instances of CRMs in the genome face a challenging problem because a majority of these interactions between transcription factors remain unknown. To investigate the reliability and accuracy of these methods, we chose twelve representative methods and applied them to predict CRMs on both the fly and human genomes. Our results show that the optimal choice of method varies depending on species and composition of the sequences in question. Different CRM representations and search strategies rely on different CRM properties, and different methods can complement one another. We provide a guide for users and key considerations for developers. We also expect that, along with new technology generating new types of genomic data, future CRM prediction methods will be able to reveal transcription binding interactions in three-dimensional space.
Thursday, December 2, 2010
[Report] Plasticity of Animal Genome Architecture Unmasked by Rapid Evolution of a Pelagic Tunicate
Authors: France Denoeud, Simon Henriet, Sutada Mungpakdee, Jean-Marc Aury, Corinne Da Silva, Henner Brinkmann, Jana Mikhaleva, Lisbeth Charlotte Olsen, Claire Jubin, Cristian Cañestro, Jean-Marie Bouquet, Gemma Danks, Julie Poulain, Coen Campsteijn, Marcin Adamski, Ismael Cross, Fekadu Yadetie, Matthieu Muffato, Alexandra Louis, Stephen Butcher, Georgia Tsagkogeorga, Anke Konrad, Sarabdeep Singh, Marit Flo Jensen, Evelyne Huynh Cong, Helen Eikeseth-Otteraa, Benjamin Noel, Véronique Anthouard, Betina M. Porcel, Rym Kachouri-Lafond, Atsuo Nishino, Matteo Ugolini, Pascal Chourrout, Hiroki Nishida, Rein Aasland, Snehalata Huzurbazar, Eric Westhof, Frédéric Delsuc, Hans Lehrach, Richard Reinhardt, Jean Weissenbach, Scott W. Roy, François Artiguenave, John H. Postlethwait, J. Robert Manak, Eric M. Thompson, Olivier Jaillon, Louis Du Pasquier, Pierre Boudinot, David A. Liberles, Jean-Nicolas Volff, Hervé Philippe, Boris Lenhard, Hugues Roest Crollius, Patrick Wincker, Daniel Chourrout"
The three-dimensional architecture of Hox cluster silencing
Spatial chromatin organization is emerging as an important mechanism to regulate the expression of genes. However, very little is known about genome architecture at high-resolution in vivo. Here, we mapped the three-dimensional organization of the human Hox clusters with chromosome conformation capture (3C) technology. We show that computational modeling of 3C data sets can identify candidate regulatory proteins of chromatin architecture and gene expression. Hox genes encode evolutionarily conserved master regulators of development which strict control has fascinated biologists for over 25 years. Proper transcriptional silencing is key to Hox function since premature expression can lead to developmental defects or human disease. We now show that the HoxA cluster is organized into multiple chromatin loops that are dependent on transcription activity. Long-range contacts were found in all four silent clusters but looping patterns were specific to each cluster. In contrast to the Drosophila homeotic bithorax complex (BX-C), we found that Polycomb proteins are only modestly required for human cluster looping and silencing. However, computational three-dimensional Hox cluster modeling identified the insulator-binding protein CTCF as a likely candidate mediating DNA loops in all clusters. Our data suggest that Hox cluster looping may represent an evolutionarily conserved structural mechanism of transcription regulation.
"Genomic repertoires of DNA-binding transcription factors across the tree of life
Sequence-specific transcription factors (TFs) are important to genetic regulation in all organisms because they recognize and directly bind to regulatory regions on DNA. Here, we survey and summarize the TF resources available. We outline the organisms for which TF annotation is provided, and discuss the criteria and methods used to annotate TFs by different databases. By using genomic TF repertoires from ~700 genomes across the tree of life, covering Bacteria, Archaea and Eukaryota, we review TF abundance with respect to the number of genes, as well as their structural complexity in diverse lineages. While typical eukaryotic TFs are longer than the average eukaryotic proteins, the inverse is true for prokaryotes. Only in eukaryotes does the same family of DNA-binding domain (DBD) occur multiple times within one polypeptide chain. This potentially increases the length and diversity of DNA-recognition sequence by reusing DBDs from the same family. We examined the increase in TF abundance with the number of genes in genomes, using the largest set of prokaryotic and eukaryotic genomes to date. As pointed out before, prokaryotic TFs increase faster than linearly. We further observe a similar relationship in eukaryotic genomes with a slower increase in TFs.
"EVOLUTION AND INHERITANCE OF EARLY EMBRYONIC PATTERNING IN D. SIMULANS AND D. SECHELLIA.
ABSTRACT
Pattern formation in Drosophila is a widely studied example of a robust developmental system. Such robust systems pose a challenge to adaptive evolution, as they mask variation which selection may otherwise act upon. Yet we find variation in the localization of expression domains (henceforth ‘stripe allometry’) in the pattern formation pathway. Specifically, we characterize differences in the gap genes giant and Kruppel, and the pair-rule gene even-skipped, which differ between the sibling species D. simulans and D. sechellia. In a double-backcross experiment, stripe allometry is consistent with maternal inheritance of stripe positioning and multiple genetic factors, with a distinct genetic basis from embryo length. Embryos produced by F1 and F2 backcross mothers exhibit novel spatial patterns of gene expression relative to the parental species, with no measurable increase in positional variance among individuals. Buffering of novel spatial patterns in the backcross genotypes suggests that robustness need not be disrupted in order for the trait to evolve, and perhaps the system is incapable of evolving to prevent the expression of all genetic variation. This limitation, and the ability of natural selection to act on minute genetic differences that are within the “margin of error” for the buffering mechanism, indicates that developmentally buffered traits can evolve without disruption of robustness
Is Transcription Factor Binding Site Turnover a Sufficient Explanation for Cis-Regulatory Sequence Divergence?
The molecular evolution of cis-regulatory sequences is not well understood. Comparisons of closely related species show that cis-regulatory sequences contain a large number of sites constrained by purifying selection. In contrast, there are a number of examples from distantly related species where cis-regulatory sequences retain little to no sequence similarity but drive similar patterns of gene expression. Binding site turnover, whereby the gain of a redundant binding site enables loss of a previously functional site, is one model by which cis-regulatory sequences can diverge without a concurrent change in function. To determine whether cis-regulatory sequence divergence is consistent with binding site turnover, we examined binding site evolution within orthologous intergenic sequences from 14 yeast species defined by their syntenic relationships with adjacent coding sequences. Both local and global alignments show that nearly all distantly related orthologous cis-regulatory sequences have no significant level of sequence similarity but are enriched for experimentally identified binding sites. Yet, a significant proportion of experimentally identified binding sites that are conserved in closely related species are absent in distantly related species and so cannot be explained by binding site turnover. Depletion of binding sites depends on the transcription factor but is detectable for a quarter of all transcription factors examined. Our results imply that binding site turnover is not a sufficient explanation for cis-regulatory sequence evolution.
"Neighbourhood Continuity Is Not Required for Correct Testis Gene Expression in Drosophila
Disrupting the linear organization of testis gene expression neighborhoods in the Drosophila genome does not affect gene expression, suggesting that neighborhood organization is not primarily driven by gene expression requirements.
Co-Evolution of Transcriptional Silencing Proteins and the DNA Elements Specifying Their Assembly
As shown by genetic assays in Saccharomyces interspecies hybrids, the co-evolution of heterochromatin assembly proteins with silencer elements allows transcriptional silencing functions to be maintained in rapidly evolving regions of the genome.