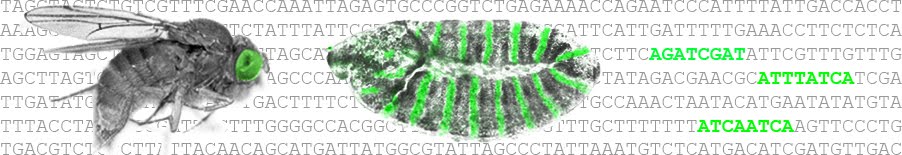
A System of Repressor Gradients Spatially Organizes the Boundaries of Bicoid-Dependent Target Genes: Hongtao Chen, Zhe Xu, Constance Mei, Danyang Yu, Stephen Small. The homeodomain (HD) protein Bicoid (Bcd) is thought to function as a gradient morphogen that positions boundaries of target genes via threshold-dependent activation mechanisms. Here, we analyze 6....
Monday, April 30, 2012
Does the Bicoid Gradient Matter?
Does the Bicoid Gradient Matter?: Siegfried Roth, Jeremy Lynch. The generation and interpretation of positional information are key processes in developmental systems. In this issue, Chen et al. report discoveries made in the Drosophila embryo that give....
Tuesday, April 24, 2012
Increasing transgene expression [Genetics]
Increasing transgene expression [Genetics]: The ability to specify the expression levels of exogenous genes inserted in the genomes of transgenic animals is critical for the success of a wide variety of experimental manipulations. Protein production can be regulated at the level of transcription, mRNA transport, mRNA half-life, or translation efficiency. In this report, we show that several well-characterized sequence elements derived from plant and insect viruses are able to function in Drosophila to increase the apparent translational efficiency of mRNAs by as much as 20-fold. These increases render expression levels sufficient for genetic constructs previously requiring multiple copies to be effective in single copy, including constructs expressing the temperature-sensitive inactivator of neuronal function Shibirets1, and for the use of cytoplasmic GFP to image the fine processes of neurons.
Friday, April 20, 2012
Mechanisms of transcriptional precision in animal development
 Mechanisms of transcriptional precision in animal development: Mounia Lagha, Jacques P. Bothma, Michael Levine. We review recently identified mechanisms of transcriptional control that ensure reliable and reproducible patterns of gene expression in natural populations of developing embryos, despite inherent....
Mechanisms of transcriptional precision in animal development: Mounia Lagha, Jacques P. Bothma, Michael Levine. We review recently identified mechanisms of transcriptional control that ensure reliable and reproducible patterns of gene expression in natural populations of developing embryos, despite inherent....
Thursday, April 19, 2012
Patterns of Cis Regulatory Variation in Diverse Human Populations
Patterns of Cis Regulatory Variation in Diverse Human Populations:
by Barbara E. Stranger, Stephen B. Montgomery, Antigone S. Dimas, Leopold Parts, Oliver Stegle, Catherine E. Ingle, Magda Sekowska, George Davey Smith, David Evans, Maria Gutierrez-Arcelus, Alkes Price, Towfique Raj, James Nisbett, Alexandra C. Nica, Claude Beazley, Richard Durbin, Panos Deloukas, Emmanouil T. Dermitzakis
The genetic basis of gene expression variation has long been studied with the aim to understand the landscape of regulatory variants, but also more recently to assist in the interpretation and elucidation of disease signals. To date, many studies have looked in specific tissues and population-based samples, but there has been limited assessment of the degree of inter-population variability in regulatory variation. We analyzed genome-wide gene expression in lymphoblastoid cell lines from a total of 726 individuals from 8 global populations from the HapMap3 project and correlated gene expression levels with HapMap3 SNPs located in cis to the genes. We describe the influence of ancestry on gene expression levels within and between these diverse human populations and uncover a non-negligible impact on global patterns of gene expression. We further dissect the specific functional pathways differentiated between populations. We also identify 5,691 expression quantitative trait loci (eQTLs) after controlling for both non-genetic factors and population admixture and observe that half of the cis-eQTLs are replicated in one or more of the populations. We highlight patterns of eQTL-sharing between populations, which are partially determined by population genetic relatedness, and discover significant sharing of eQTL effects between Asians, European-admixed, and African subpopulations. Specifically, we observe that both the effect size and the direction of effect for eQTLs are highly conserved across populations. We observe an increasing proximity of eQTLs toward the transcription start site as sharing of eQTLs among populations increases, highlighting that variants close to TSS have stronger effects and therefore are more likely to be detected across a wider panel of populations. Together these results offer a unique picture and resource of the degree of differentiation among human populations in functional regulatory variation and provide an estimate for the transferability of complex trait variants across populations.
by Barbara E. Stranger, Stephen B. Montgomery, Antigone S. Dimas, Leopold Parts, Oliver Stegle, Catherine E. Ingle, Magda Sekowska, George Davey Smith, David Evans, Maria Gutierrez-Arcelus, Alkes Price, Towfique Raj, James Nisbett, Alexandra C. Nica, Claude Beazley, Richard Durbin, Panos Deloukas, Emmanouil T. Dermitzakis
The genetic basis of gene expression variation has long been studied with the aim to understand the landscape of regulatory variants, but also more recently to assist in the interpretation and elucidation of disease signals. To date, many studies have looked in specific tissues and population-based samples, but there has been limited assessment of the degree of inter-population variability in regulatory variation. We analyzed genome-wide gene expression in lymphoblastoid cell lines from a total of 726 individuals from 8 global populations from the HapMap3 project and correlated gene expression levels with HapMap3 SNPs located in cis to the genes. We describe the influence of ancestry on gene expression levels within and between these diverse human populations and uncover a non-negligible impact on global patterns of gene expression. We further dissect the specific functional pathways differentiated between populations. We also identify 5,691 expression quantitative trait loci (eQTLs) after controlling for both non-genetic factors and population admixture and observe that half of the cis-eQTLs are replicated in one or more of the populations. We highlight patterns of eQTL-sharing between populations, which are partially determined by population genetic relatedness, and discover significant sharing of eQTL effects between Asians, European-admixed, and African subpopulations. Specifically, we observe that both the effect size and the direction of effect for eQTLs are highly conserved across populations. We observe an increasing proximity of eQTLs toward the transcription start site as sharing of eQTLs among populations increases, highlighting that variants close to TSS have stronger effects and therefore are more likely to be detected across a wider panel of populations. Together these results offer a unique picture and resource of the degree of differentiation among human populations in functional regulatory variation and provide an estimate for the transferability of complex trait variants across populations.
[Report] A Segmentation Clock with Two-Segment Periodicity in Insects
[Report] A Segmentation Clock with Two-Segment Periodicity in Insects: Oscillating gene expression, a key feature of vertebrate segmentation, is shown to occur during segmentation in beetles.
Authors: Andres F. Sarrazin, Andrew D. Peel, Michalis Averof
Authors: Andres F. Sarrazin, Andrew D. Peel, Michalis Averof
Saturday, April 14, 2012
Large-Scale Cellular-Resolution Gene Profiling in Human Neocortex Reveals Species-Specific Molecular Signatures
Large-Scale Cellular-Resolution Gene Profiling in Human Neocortex Reveals Species-Specific Molecular Signatures: Hongkui Zeng, Elaine H. Shen, John G. Hohmann, Seung Wook Oh, Amy Bernard, Joshua J. Royall, Katie J. Glattfelder, Susan M. Sunkin, John A. Morris, Angela L. Guillozet-Bongaarts, Kimberly A. Smith, Amanda J. Ebbert, Beryl Swanson, Leonard Kuan, Damon T. Page, Caroline C. Overly, Ed S. Lein, Michael J. Hawrylycz, Patrick R. Hof, Thomas M. Hyde, Joel E. Kleinman, Allan R. Jones.
Although there have been major advances in elucidating the functional biology of the human brain, relatively little is known of its cellular and molecular organization. Here we report a large-scal....
Although there have been major advances in elucidating the functional biology of the human brain, relatively little is known of its cellular and molecular organization. Here we report a large-scal....
Dynamic Transformations of Genome-wide Epigenetic Marking and Transcriptional Control Establish T Cell Identity
Dynamic Transformations of Genome-wide Epigenetic Marking and Transcriptional Control Establish T Cell Identity: Jingli A. Zhang, Ali Mortazavi, Brian A. Williams, Barbara J. Wold, Ellen V. Rothenberg.
T cell development comprises a stepwise process of commitment from a multipotent precursor. To define molecular mechanisms controlling this progression, we probed five stages spanning the commitme....
T cell development comprises a stepwise process of commitment from a multipotent precursor. To define molecular mechanisms controlling this progression, we probed five stages spanning the commitme....
Thursday, April 12, 2012
Genome-wide protein–DNA binding dynamics suggest a molecular clutch for transcription factor function
Genome-wide protein–DNA binding dynamics suggest a molecular clutch for transcription factor function:
Genome-wide protein–DNA binding dynamics suggest a molecular clutch for transcription factor function
Nature 484, 7393 (2012). doi:10.1038/nature10985
Authors: Colin R. Lickwar, Florian Mueller, Sean E. Hanlon, James G. McNally & Jason D. Lieb
Dynamic access to genetic information is central to organismal development and environmental response. Consequently, genomic processes must be regulated by mechanisms that alter genome function relatively rapidly. Conventional chromatin immunoprecipitation (ChIP) experiments measure transcription factor occupancy, but give no indication of kinetics and are poor predictors of transcription factor function at a given locus. To measure transcription-factor-binding dynamics across the genome, we performed competition ChIP (refs 6, 7) with a sequence-specific Saccharomyces cerevisiae transcription factor, Rap1 (ref. 8). Rap1-binding dynamics and Rap1 occupancy were only weakly correlated (R2 = 0.14), but binding dynamics were more strongly linked to function than occupancy. Long Rap1 residence was coupled to transcriptional activation, whereas fast binding turnover, which we refer to as ‘treadmilling’, was linked to low transcriptional output. Thus, DNA-binding events that seem identical by conventional ChIP may have different underlying modes of interaction that lead to opposing functional outcomes. We propose that transcription factor binding turnover is a major point of regulation in determining the functional consequences of transcription factor binding, and is mediated mainly by control of competition between transcription factors and nucleosomes. Our model predicts a clutch-like mechanism that rapidly engages a treadmilling transcription factor into a stable binding state, or vice versa, to modulate transcription factor function.

Genome-wide protein–DNA binding dynamics suggest a molecular clutch for transcription factor function
Nature 484, 7393 (2012). doi:10.1038/nature10985
Authors: Colin R. Lickwar, Florian Mueller, Sean E. Hanlon, James G. McNally & Jason D. Lieb
Dynamic access to genetic information is central to organismal development and environmental response. Consequently, genomic processes must be regulated by mechanisms that alter genome function relatively rapidly. Conventional chromatin immunoprecipitation (ChIP) experiments measure transcription factor occupancy, but give no indication of kinetics and are poor predictors of transcription factor function at a given locus. To measure transcription-factor-binding dynamics across the genome, we performed competition ChIP (refs 6, 7) with a sequence-specific Saccharomyces cerevisiae transcription factor, Rap1 (ref. 8). Rap1-binding dynamics and Rap1 occupancy were only weakly correlated (R2 = 0.14), but binding dynamics were more strongly linked to function than occupancy. Long Rap1 residence was coupled to transcriptional activation, whereas fast binding turnover, which we refer to as ‘treadmilling’, was linked to low transcriptional output. Thus, DNA-binding events that seem identical by conventional ChIP may have different underlying modes of interaction that lead to opposing functional outcomes. We propose that transcription factor binding turnover is a major point of regulation in determining the functional consequences of transcription factor binding, and is mediated mainly by control of competition between transcription factors and nucleosomes. Our model predicts a clutch-like mechanism that rapidly engages a treadmilling transcription factor into a stable binding state, or vice versa, to modulate transcription factor function.
Monday, April 9, 2012
Genomic repertoires of DNA-binding transcription factors across the tree of life
Genomic repertoires of DNA-binding transcription factors across the tree of life:
Sequence-specific transcription factors (TFs) are important to genetic regulation in all organisms because they recognize and directly bind to regulatory regions on DNA. Here, we survey and summarize the TF resources available. We outline the organisms for which TF annotation is provided, and discuss the criteria and methods used to annotate TFs by different databases. By using genomic TF repertoires from ~700 genomes across the tree of life, covering Bacteria, Archaea and Eukaryota, we review TF abundance with respect to the number of genes, as well as their structural complexity in diverse lineages. While typical eukaryotic TFs are longer than the average eukaryotic proteins, the inverse is true for prokaryotes. Only in eukaryotes does the same family of DNA-binding domain (DBD) occur multiple times within one polypeptide chain. This potentially increases the length and diversity of DNA-recognition sequence by reusing DBDs from the same family. We examined the increase in TF abundance with the number of genes in genomes, using the largest set of prokaryotic and eukaryotic genomes to date. As pointed out before, prokaryotic TFs increase faster than linearly. We further observe a similar relationship in eukaryotic genomes with a slower increase in TFs.
Sequence-specific transcription factors (TFs) are important to genetic regulation in all organisms because they recognize and directly bind to regulatory regions on DNA. Here, we survey and summarize the TF resources available. We outline the organisms for which TF annotation is provided, and discuss the criteria and methods used to annotate TFs by different databases. By using genomic TF repertoires from ~700 genomes across the tree of life, covering Bacteria, Archaea and Eukaryota, we review TF abundance with respect to the number of genes, as well as their structural complexity in diverse lineages. While typical eukaryotic TFs are longer than the average eukaryotic proteins, the inverse is true for prokaryotes. Only in eukaryotes does the same family of DNA-binding domain (DBD) occur multiple times within one polypeptide chain. This potentially increases the length and diversity of DNA-recognition sequence by reusing DBDs from the same family. We examined the increase in TF abundance with the number of genes in genomes, using the largest set of prokaryotic and eukaryotic genomes to date. As pointed out before, prokaryotic TFs increase faster than linearly. We further observe a similar relationship in eukaryotic genomes with a slower increase in TFs.
The three-dimensional architecture of Hox cluster silencing
The three-dimensional architecture of Hox cluster silencing:
Spatial chromatin organization is emerging as an important mechanism to regulate the expression of genes. However, very little is known about genome architecture at high-resolution in vivo. Here, we mapped the three-dimensional organization of the human Hox clusters with chromosome conformation capture (3C) technology. We show that computational modeling of 3C data sets can identify candidate regulatory proteins of chromatin architecture and gene expression. Hox genes encode evolutionarily conserved master regulators of development which strict control has fascinated biologists for over 25 years. Proper transcriptional silencing is key to Hox function since premature expression can lead to developmental defects or human disease. We now show that the HoxA cluster is organized into multiple chromatin loops that are dependent on transcription activity. Long-range contacts were found in all four silent clusters but looping patterns were specific to each cluster. In contrast to the Drosophila homeotic bithorax complex (BX-C), we found that Polycomb proteins are only modestly required for human cluster looping and silencing. However, computational three-dimensional Hox cluster modeling identified the insulator-binding protein CTCF as a likely candidate mediating DNA loops in all clusters. Our data suggest that Hox cluster looping may represent an evolutionarily conserved structural mechanism of transcription regulation.
Spatial chromatin organization is emerging as an important mechanism to regulate the expression of genes. However, very little is known about genome architecture at high-resolution in vivo. Here, we mapped the three-dimensional organization of the human Hox clusters with chromosome conformation capture (3C) technology. We show that computational modeling of 3C data sets can identify candidate regulatory proteins of chromatin architecture and gene expression. Hox genes encode evolutionarily conserved master regulators of development which strict control has fascinated biologists for over 25 years. Proper transcriptional silencing is key to Hox function since premature expression can lead to developmental defects or human disease. We now show that the HoxA cluster is organized into multiple chromatin loops that are dependent on transcription activity. Long-range contacts were found in all four silent clusters but looping patterns were specific to each cluster. In contrast to the Drosophila homeotic bithorax complex (BX-C), we found that Polycomb proteins are only modestly required for human cluster looping and silencing. However, computational three-dimensional Hox cluster modeling identified the insulator-binding protein CTCF as a likely candidate mediating DNA loops in all clusters. Our data suggest that Hox cluster looping may represent an evolutionarily conserved structural mechanism of transcription regulation.
The role of transcription factories-mediated interchromosomal contacts in the organization of nuclear architecture
The role of transcription factories-mediated interchromosomal contacts in the organization of nuclear architecture:
Using numerical simulations, we investigate the underlying physical effects responsible for the overall organization of chromosomal territories in interphase nuclei. In particular, we address the following three questions: (i) why are chromosomal territories with relatively high transcriptional activity on average, closer to the centre of cell's nucleus than those with the lower activity? (ii) Why are actively transcribed genes usually located at the periphery of their chromosomal territories? (iii) Why are pair-wise contacts between active and inactive genes less frequent than those involving only active or only inactive genes? We show that transcription factories-mediated contacts between active genes belonging to different chromosomal territories are instrumental for all these features of nuclear organization to emerge spontaneously due to entropic effects arising when chromatin fibres are highly crowded.
Using numerical simulations, we investigate the underlying physical effects responsible for the overall organization of chromosomal territories in interphase nuclei. In particular, we address the following three questions: (i) why are chromosomal territories with relatively high transcriptional activity on average, closer to the centre of cell's nucleus than those with the lower activity? (ii) Why are actively transcribed genes usually located at the periphery of their chromosomal territories? (iii) Why are pair-wise contacts between active and inactive genes less frequent than those involving only active or only inactive genes? We show that transcription factories-mediated contacts between active genes belonging to different chromosomal territories are instrumental for all these features of nuclear organization to emerge spontaneously due to entropic effects arising when chromatin fibres are highly crowded.
Thursday, April 5, 2012
Pathway analysis of genomic data: concepts, methods and prospects for future development
Pathway analysis of genomic data: concepts, methods and prospects for future development: Vijay K. Ramanan, Li Shen, Jason H. Moore, Andrew J. Saykin. Genome-wide data sets are increasingly being used to identify biological pathways and networks underlying complex diseases. In particular, analyzing genomic data through sets defined by functional....
The genomic basis of adaptive evolution in threespine sticklebacks
The genomic basis of adaptive evolution in threespine sticklebacks:
The genomic basis of adaptive evolution in threespine sticklebacks
Nature 484, 7392 (2012). doi:10.1038/nature10944
Authors: Felicity C. Jones, Manfred G. Grabherr, Yingguang Frank Chan, Pamela Russell, Evan Mauceli, Jeremy Johnson, Ross Swofford, Mono Pirun, Michael C. Zody, Simon White, Ewan Birney, Stephen Searle, Jeremy Schmutz, Jane Grimwood, Mark C. Dickson, Richard M. Myers, Craig T. Miller, Brian R. Summers, Anne K. Knecht, Shannon D. Brady, Haili Zhang, Alex A. Pollen, Timothy Howes, Chris Amemiya, Eric S. Lander, Federica Di Palma, Kerstin Lindblad-Toh & David M. Kingsley
Marine stickleback fish have colonized and adapted to thousands of streams and lakes formed since the last ice age, providing an exceptional opportunity to characterize genomic mechanisms underlying repeated ecological adaptation in nature. Here we develop a high-quality reference genome assembly for threespine sticklebacks. By

The genomic basis of adaptive evolution in threespine sticklebacks
Nature 484, 7392 (2012). doi:10.1038/nature10944
Authors: Felicity C. Jones, Manfred G. Grabherr, Yingguang Frank Chan, Pamela Russell, Evan Mauceli, Jeremy Johnson, Ross Swofford, Mono Pirun, Michael C. Zody, Simon White, Ewan Birney, Stephen Searle, Jeremy Schmutz, Jane Grimwood, Mark C. Dickson, Richard M. Myers, Craig T. Miller, Brian R. Summers, Anne K. Knecht, Shannon D. Brady, Haili Zhang, Alex A. Pollen, Timothy Howes, Chris Amemiya, Eric S. Lander, Federica Di Palma, Kerstin Lindblad-Toh & David M. Kingsley
Marine stickleback fish have colonized and adapted to thousands of streams and lakes formed since the last ice age, providing an exceptional opportunity to characterize genomic mechanisms underlying repeated ecological adaptation in nature. Here we develop a high-quality reference genome assembly for threespine sticklebacks. By
Wednesday, April 4, 2012
A gigantic feathered dinosaur from the Lower Cretaceous of China
A gigantic feathered dinosaur from the Lower Cretaceous of China:
A gigantic feathered dinosaur from the Lower Cretaceous of China
Nature 484, 7392 (2012). doi:10.1038/nature10906
Authors: Xing Xu, Kebai Wang, Ke Zhang, Qingyu Ma, Lida Xing, Corwin Sullivan, Dongyu Hu, Shuqing Cheng & Shuo Wang
Numerous feathered dinosaur specimens have recently been recovered from the Middle–Upper Jurassic and Lower Cretaceous deposits of northeastern China, but most of them represent small animals. Here we report the discovery of a gigantic new basal tyrannosauroid, Yutyrannus huali gen. et sp. nov., based on three nearly complete skeletons representing two distinct ontogenetic stages from the Lower Cretaceous Yixian Formation of Liaoning Province, China. Y. huali shares some features, particularly of the cranium, with derived tyrannosauroids, but is similar to other basal tyrannosauroids in possessing a three-fingered manus and a typical theropod pes. Morphometric analysis suggests that Y. huali differed from tyrannosaurids in its growth strategy. Most significantly, Y. huali bears long filamentous feathers, thus providing direct evidence for the presence of extensively feathered gigantic dinosaurs and offering new insights into early feather evolution.

A gigantic feathered dinosaur from the Lower Cretaceous of China
Nature 484, 7392 (2012). doi:10.1038/nature10906
Authors: Xing Xu, Kebai Wang, Ke Zhang, Qingyu Ma, Lida Xing, Corwin Sullivan, Dongyu Hu, Shuqing Cheng & Shuo Wang
Numerous feathered dinosaur specimens have recently been recovered from the Middle–Upper Jurassic and Lower Cretaceous deposits of northeastern China, but most of them represent small animals. Here we report the discovery of a gigantic new basal tyrannosauroid, Yutyrannus huali gen. et sp. nov., based on three nearly complete skeletons representing two distinct ontogenetic stages from the Lower Cretaceous Yixian Formation of Liaoning Province, China. Y. huali shares some features, particularly of the cranium, with derived tyrannosauroids, but is similar to other basal tyrannosauroids in possessing a three-fingered manus and a typical theropod pes. Morphometric analysis suggests that Y. huali differed from tyrannosaurids in its growth strategy. Most significantly, Y. huali bears long filamentous feathers, thus providing direct evidence for the presence of extensively feathered gigantic dinosaurs and offering new insights into early feather evolution.
Tuesday, April 3, 2012
Biphasic wnt8a expression is achieved through interactions of multiple regulatory inputs
Biphasic wnt8a expression is achieved through interactions of multiple regulatory inputs:
Abstract
Background:
Vertebrate axis development depends upon wnt8a transcription in a dynamic pool of mesoderm progenitors at the posterior pole of the gastrulating embryo. The transcriptional mechanisms controlling wnt8a expression are not understood, but previous studies identified two phases of wnt8a expression in zebrafish: Nodal-dependent activation during early gastrulation (phase I) and No tail (Ntl)-dependent regulation from mid gastrula stages (phase II).
Results:
We identified two upstream cis-regulatory regions, proximal and distal, each of which possesses a promoter. The proximal regulatory region contains a margin-specific enhancer that is required for both the Nodal and Ntl responses. Phase I expression requires Nodal activation of the margin enhancer in combination with the transcription factor Zbtb4 and the distal regulatory region. Phase II expression requires Ntl regulation of the margin enhancer in the context of the proximal regulatory region. An additional mechanism is required to ensure the transition from phase I to phase II regulation. Analysis of stickleback wnt8a suggests this mechanism of regulation may be conserved.
Conclusion:
The seemingly simple wnt8a expression pattern reflects complex interactions of multiple regulatory inputs. Developmental Dynamics, 2012. © 2012 Wiley Periodicals, Inc.
Chromatin control of pancreatic transcription [Genetics]
Chromatin control of pancreatic transcription [Genetics]: Under the instruction of cell-fate–determining, DNA-binding transcription factors, chromatin-modifying enzymes mediate and maintain cell states throughout development in multicellular organisms. Currently, small molecules modulating the activity of several classes of chromatin-modifying enzymes are available, including clinically approved histone deacetylase (HDAC) and DNA methyltransferase (DNMT) inhibitors. We describe the genome-wide expression changes induced by 29 compounds targeting HDACs, DNMTs, histone lysine methyltransferases (HKMTs), and protein arginine methyltransferases (PRMTs) in pancreatic α- and β-cell lines. HDAC inhibitors regulate several hundred transcripts irrespective of the cell type, with distinct clusters of dissimilar activity for hydroxamic acids and orthoamino anilides. In contrast, compounds targeting histone methyltransferases modulate the expression of restricted gene sets in distinct cell types. For example, we find that G9a/GLP methyltransferase inhibitors selectively up-regulate the cholesterol biosynthetic pathway in pancreatic but not liver cells. These data suggest that, despite their conservation across the entire genome and in different cell types, chromatin pathways can be targeted to modulate the expression of selected transcripts.
Integration of an abdominal Hox complex with Pax2 yields cell-specific EGF secretion from Drosophila sensory precursor cells [RESEARCH ARTICLES]
Integration of an abdominal Hox complex with Pax2 yields cell-specific EGF secretion from Drosophila sensory precursor cells [RESEARCH ARTICLES]: David Li-Kroeger, Tiffany A. Cook, and Brian Gebelein
Cis-regulatory modules (CRMs) ensure specific developmental outcomes by mediating both proper spatiotemporal gene expression patterns and appropriate transcriptional levels. In Drosophila, the precise transcriptional control of the serine protease rhomboid regulates EGF signaling to specify distinct cell types. Recently, we identified a CRM that activates rhomboid expression and thereby EGF secretion from a subset of abdominal sensory organ precursor cells (SOPs) to induce an appropriate number of lipid-processing cells called oenocytes. Here, we use scanning mutagenesis coupled with reporter assays, biochemistry and genetics to dissect the transcriptional mechanisms regulating SOP-specific rhomboid activation. Our results show that proper spatial activity of the rhomboid CRM is dependent upon direct integration of the abdomen-specific Hox factor Abdominal-A and the SOP-restricted Pax2 factor. In addition, we show that the Extradenticle and Homothorax Hox co-factors are differentially integrated on the rhomboid CRM by abdominal versus thoracic Hox proteins in the presence of Pax2. Last, we show that Abdominal-A uses both Pax2-dependent and Pax2-independent mechanisms to stimulate rhomboid CRM activity to induce proper oenocyte numbers. Thus, these data demonstrate how a CRM integrates Hox and neural transcriptional inputs to regulate the appropriate spatial pattern and levels of EGF secretion to specify an essential cell fate.
Cis-regulatory modules (CRMs) ensure specific developmental outcomes by mediating both proper spatiotemporal gene expression patterns and appropriate transcriptional levels. In Drosophila, the precise transcriptional control of the serine protease rhomboid regulates EGF signaling to specify distinct cell types. Recently, we identified a CRM that activates rhomboid expression and thereby EGF secretion from a subset of abdominal sensory organ precursor cells (SOPs) to induce an appropriate number of lipid-processing cells called oenocytes. Here, we use scanning mutagenesis coupled with reporter assays, biochemistry and genetics to dissect the transcriptional mechanisms regulating SOP-specific rhomboid activation. Our results show that proper spatial activity of the rhomboid CRM is dependent upon direct integration of the abdomen-specific Hox factor Abdominal-A and the SOP-restricted Pax2 factor. In addition, we show that the Extradenticle and Homothorax Hox co-factors are differentially integrated on the rhomboid CRM by abdominal versus thoracic Hox proteins in the presence of Pax2. Last, we show that Abdominal-A uses both Pax2-dependent and Pax2-independent mechanisms to stimulate rhomboid CRM activity to induce proper oenocyte numbers. Thus, these data demonstrate how a CRM integrates Hox and neural transcriptional inputs to regulate the appropriate spatial pattern and levels of EGF secretion to specify an essential cell fate.
Dynamic in vivo binding of transcription factors to cis-regulatory modules of cer and gsc in the stepwise formation of the Spemann-Mangold organizer [RESEARCH ARTICLES]
Dynamic in vivo binding of transcription factors to cis-regulatory modules of cer and gsc in the stepwise formation of the Spemann-Mangold organizer [RESEARCH ARTICLES]: Norihiro Sudou, Shinji Yamamoto, Hajime Ogino, and Masanori Taira
How multiple developmental cues are integrated on cis-regulatory modules (CRMs) for cell fate decisions remains uncertain. The Spemann–Mangold organizer in Xenopus embryos expresses the transcription factors Lim1/Lhx1, Otx2, Mix1, Siamois (Sia) and VegT. Reporter analyses using sperm nuclear transplantation and DNA injection showed that cerberus (cer) and goosecoid (gsc) are activated by the aforementioned transcription factors through CRMs conserved between X. laevis and X. tropicalis. ChIP-qPCR analysis for the five transcription factors revealed that cer and gsc CRMs are initially bound by both Sia and VegT at the late blastula stage, and subsequently bound by all five factors at the gastrula stage. At the neurula stage, only binding of Lim1 and Otx2 to the gsc CRM, among others, persists, which corresponds to their co-expression in the prechordal plate. Based on these data, together with detailed expression pattern analysis, we propose a new model of stepwise formation of the organizer, in which (1) maternal VegT and Wnt-induced Sia first bind to CRMs at the blastula stage; then (2) Nodal-inducible Lim1, Otx2, Mix1 and zygotic VegT are bound to CRMs in the dorsal endodermal and mesodermal regions where all these genes are co-expressed; and (3) these two regions are combined at the gastrula stage to form the organizer. Thus, the in vivo dynamics of multiple transcription factors highlight their roles in the initiation and maintenance of gene expression, and also reveal the stepwise integration of maternal, Nodal and Wnt signaling on CRMs of organizer genes to generate the organizer.
How multiple developmental cues are integrated on cis-regulatory modules (CRMs) for cell fate decisions remains uncertain. The Spemann–Mangold organizer in Xenopus embryos expresses the transcription factors Lim1/Lhx1, Otx2, Mix1, Siamois (Sia) and VegT. Reporter analyses using sperm nuclear transplantation and DNA injection showed that cerberus (cer) and goosecoid (gsc) are activated by the aforementioned transcription factors through CRMs conserved between X. laevis and X. tropicalis. ChIP-qPCR analysis for the five transcription factors revealed that cer and gsc CRMs are initially bound by both Sia and VegT at the late blastula stage, and subsequently bound by all five factors at the gastrula stage. At the neurula stage, only binding of Lim1 and Otx2 to the gsc CRM, among others, persists, which corresponds to their co-expression in the prechordal plate. Based on these data, together with detailed expression pattern analysis, we propose a new model of stepwise formation of the organizer, in which (1) maternal VegT and Wnt-induced Sia first bind to CRMs at the blastula stage; then (2) Nodal-inducible Lim1, Otx2, Mix1 and zygotic VegT are bound to CRMs in the dorsal endodermal and mesodermal regions where all these genes are co-expressed; and (3) these two regions are combined at the gastrula stage to form the organizer. Thus, the in vivo dynamics of multiple transcription factors highlight their roles in the initiation and maintenance of gene expression, and also reveal the stepwise integration of maternal, Nodal and Wnt signaling on CRMs of organizer genes to generate the organizer.
Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex [REVIEW]
Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex [REVIEW]: Kenneth Y. Kwan, Nenad Sestan, and E. S. Anton
The cerebral neocortex is segregated into six horizontal layers, each containing unique populations of molecularly and functionally distinct excitatory projection (pyramidal) neurons and inhibitory interneurons. Development of the neocortex requires the orchestrated execution of a series of crucial processes, including the migration of young neurons into appropriate positions within the nascent neocortex, and the acquisition of layer-specific neuronal identities and axonal projections. Here, we discuss emerging evidence supporting the notion that the migration and final laminar positioning of cortical neurons are also co-regulated by cell type- and layer-specific transcription factors that play concomitant roles in determining the molecular identity and axonal connectivity of these neurons. These transcriptional programs thus provide direct links between the mechanisms controlling the laminar position and identity of cortical neurons.
The cerebral neocortex is segregated into six horizontal layers, each containing unique populations of molecularly and functionally distinct excitatory projection (pyramidal) neurons and inhibitory interneurons. Development of the neocortex requires the orchestrated execution of a series of crucial processes, including the migration of young neurons into appropriate positions within the nascent neocortex, and the acquisition of layer-specific neuronal identities and axonal projections. Here, we discuss emerging evidence supporting the notion that the migration and final laminar positioning of cortical neurons are also co-regulated by cell type- and layer-specific transcription factors that play concomitant roles in determining the molecular identity and axonal connectivity of these neurons. These transcriptional programs thus provide direct links between the mechanisms controlling the laminar position and identity of cortical neurons.
Monday, April 2, 2012
Cell-type-specific nuclei purification from whole animals for genome-wide expression and chromatin profiling [METHOD]
Cell-type-specific nuclei purification from whole animals for genome-wide expression and chromatin profiling [METHOD]:
An understanding of developmental processes requires knowledge of transcriptional and epigenetic landscapes at the level of tissues and ultimately individual cells. However, obtaining tissue- or cell-type-specific expression and chromatin profiles for animals has been challenging. Here we describe a method for purifying nuclei from specific cell types of animal models that allows simultaneous determination of both expression and chromatin profiles. The method is based on in vivo biotin-labeling of the nuclear envelope and subsequent affinity purification of nuclei. We describe the use of the method to isolate nuclei from muscle of adult Caenorhabditis elegans and from mesoderm of Drosophila melanogaster embryos. As a case study, we determined expression and nucleosome occupancy profiles for affinity-purified nuclei from C. elegans muscle. We identified hundreds of genes that are specifically expressed in muscle tissues and found that these genes are depleted of nucleosomes at promoters and gene bodies in muscle relative to other tissues. This method should be universally applicable to all model systems that allow transgenesis and will make it possible to determine epigenetic and expression profiles of different tissues and cell types.
An understanding of developmental processes requires knowledge of transcriptional and epigenetic landscapes at the level of tissues and ultimately individual cells. However, obtaining tissue- or cell-type-specific expression and chromatin profiles for animals has been challenging. Here we describe a method for purifying nuclei from specific cell types of animal models that allows simultaneous determination of both expression and chromatin profiles. The method is based on in vivo biotin-labeling of the nuclear envelope and subsequent affinity purification of nuclei. We describe the use of the method to isolate nuclei from muscle of adult Caenorhabditis elegans and from mesoderm of Drosophila melanogaster embryos. As a case study, we determined expression and nucleosome occupancy profiles for affinity-purified nuclei from C. elegans muscle. We identified hundreds of genes that are specifically expressed in muscle tissues and found that these genes are depleted of nucleosomes at promoters and gene bodies in muscle relative to other tissues. This method should be universally applicable to all model systems that allow transgenesis and will make it possible to determine epigenetic and expression profiles of different tissues and cell types.
Evolutionary rate covariation reveals shared functionality and coexpression of genes [RESEARCH]
Evolutionary rate covariation reveals shared functionality and coexpression of genes [RESEARCH]:
Evolutionary rate covariation (ERC) is a phylogenetic signature that reflects the covariation of a pair of proteins over evolutionary time. ERC is typically elevated between interacting proteins and so is a promising signature to characterize molecular and functional interactions across the genome. ERC is often assumed to result from compensatory changes at interaction interfaces (i.e., intermolecular coevolution); however, its origin is still unclear and is likely to be complex. Here, we determine the biological factors responsible for ERC in a proteome-wide data set of 4459 proteins in 18 budding yeast species. We show that direct physical interaction is not required to produce ERC, because we observe strong correlations between noninteracting but cofunctional enzymes. We also demonstrate that ERC is uniformly distributed along the protein primary sequence, suggesting that intermolecular coevolution is not generally responsible for ERC between physically interacting proteins. Using multivariate analysis, we show that a pair of proteins is likely to exhibit ERC if they share a biological function or if their expression levels coevolve between species. Thus, ERC indicates shared function and coexpression of protein pairs and not necessarily coevolution between sites, as has been assumed in previous studies. This full interpretation of ERC now provides us with a powerful tool to assign uncharacterized proteins to functional groups and to determine the interconnectedness between entire genetic pathways.
Evolutionary rate covariation (ERC) is a phylogenetic signature that reflects the covariation of a pair of proteins over evolutionary time. ERC is typically elevated between interacting proteins and so is a promising signature to characterize molecular and functional interactions across the genome. ERC is often assumed to result from compensatory changes at interaction interfaces (i.e., intermolecular coevolution); however, its origin is still unclear and is likely to be complex. Here, we determine the biological factors responsible for ERC in a proteome-wide data set of 4459 proteins in 18 budding yeast species. We show that direct physical interaction is not required to produce ERC, because we observe strong correlations between noninteracting but cofunctional enzymes. We also demonstrate that ERC is uniformly distributed along the protein primary sequence, suggesting that intermolecular coevolution is not generally responsible for ERC between physically interacting proteins. Using multivariate analysis, we show that a pair of proteins is likely to exhibit ERC if they share a biological function or if their expression levels coevolve between species. Thus, ERC indicates shared function and coexpression of protein pairs and not necessarily coevolution between sites, as has been assumed in previous studies. This full interpretation of ERC now provides us with a powerful tool to assign uncharacterized proteins to functional groups and to determine the interconnectedness between entire genetic pathways.
The TAGteam motif facilitates binding of 21 sequence-specific transcription factors in the Drosophila embryo [RESEARCH]
The TAGteam motif facilitates binding of 21 sequence-specific transcription factors in the Drosophila embryo [RESEARCH]:
Highly overlapping patterns of genome-wide binding of many distinct transcription factors have been observed in worms, insects, and mammals, but the origins and consequences of this overlapping binding remain unclear. While analyzing chromatin immunoprecipitation data sets from 21 sequence-specific transcription factors active in the Drosophila embryo, we found that binding of all factors exhibits a dose-dependent relationship with "TAGteam" sequence motifs bound by the zinc finger protein Vielfaltig, also known as Zelda, a recently discovered activator of the zygotic genome. TAGteam motifs are present and well conserved in highly bound regions, and are associated with transcription factor binding even in the absence of canonical recognition motifs for these factors. Furthermore, levels of binding in promoters and enhancers of zygotically transcribed genes are correlated with RNA polymerase II occupancy and gene expression levels. Our results suggest that Vielfaltig acts as a master regulator of early development by facilitating the genome-wide establishment of overlapping patterns of binding of diverse transcription factors that drive global gene expression.
Highly overlapping patterns of genome-wide binding of many distinct transcription factors have been observed in worms, insects, and mammals, but the origins and consequences of this overlapping binding remain unclear. While analyzing chromatin immunoprecipitation data sets from 21 sequence-specific transcription factors active in the Drosophila embryo, we found that binding of all factors exhibits a dose-dependent relationship with "TAGteam" sequence motifs bound by the zinc finger protein Vielfaltig, also known as Zelda, a recently discovered activator of the zygotic genome. TAGteam motifs are present and well conserved in highly bound regions, and are associated with transcription factor binding even in the absence of canonical recognition motifs for these factors. Furthermore, levels of binding in promoters and enhancers of zygotically transcribed genes are correlated with RNA polymerase II occupancy and gene expression levels. Our results suggest that Vielfaltig acts as a master regulator of early development by facilitating the genome-wide establishment of overlapping patterns of binding of diverse transcription factors that drive global gene expression.
An ancient genomic regulatory block conserved across bilaterians and its dismantling in tetrapods by retrogene replacement [RESEARCH]
An ancient genomic regulatory block conserved across bilaterians and its dismantling in tetrapods by retrogene replacement [RESEARCH]:
Developmental genes are regulated by complex, distantly located cis-regulatory modules (CRMs), often forming genomic regulatory blocks (GRBs) that are conserved among vertebrates and among insects. We have investigated GRBs associated with Iroquois homeobox genes in 39 metazoans. Despite 600 million years of independent evolution, Iroquois genes are linked to ankyrin-repeat-containing Sowah genes in nearly all studied bilaterians. We show that Iroquois-specific CRMs populate the Sowah locus, suggesting that regulatory constraints underlie the maintenance of the Iroquois–Sowah syntenic block. Surprisingly, tetrapod Sowah orthologs are intronless and not associated with Iroquois; however, teleost and elephant shark data demonstrate that this is a derived feature, and that many Iroquois–CRMs were ancestrally located within Sowah introns. Retroposition, gene, and genome duplication have allowed selective elimination of Sowah exons from the Iroquois regulatory landscape while keeping associated CRMs, resulting in large associated gene deserts. These results highlight the importance of CRMs in imposing constraints to genome architecture, even across large phylogenetic distances, and of gene duplication-mediated genetic redundancy to disentangle these constraints, increasing genomic plasticity.
Developmental genes are regulated by complex, distantly located cis-regulatory modules (CRMs), often forming genomic regulatory blocks (GRBs) that are conserved among vertebrates and among insects. We have investigated GRBs associated with Iroquois homeobox genes in 39 metazoans. Despite 600 million years of independent evolution, Iroquois genes are linked to ankyrin-repeat-containing Sowah genes in nearly all studied bilaterians. We show that Iroquois-specific CRMs populate the Sowah locus, suggesting that regulatory constraints underlie the maintenance of the Iroquois–Sowah syntenic block. Surprisingly, tetrapod Sowah orthologs are intronless and not associated with Iroquois; however, teleost and elephant shark data demonstrate that this is a derived feature, and that many Iroquois–CRMs were ancestrally located within Sowah introns. Retroposition, gene, and genome duplication have allowed selective elimination of Sowah exons from the Iroquois regulatory landscape while keeping associated CRMs, resulting in large associated gene deserts. These results highlight the importance of CRMs in imposing constraints to genome architecture, even across large phylogenetic distances, and of gene duplication-mediated genetic redundancy to disentangle these constraints, increasing genomic plasticity.
Subscribe to:
Posts (Atom)