Wednesday, January 30, 2013
Creating gradients by morphogen shuttling
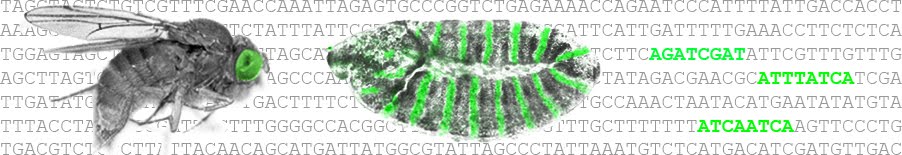
Creating gradients by morphogen shuttling: Ben-Zion Shilo, Michal Haskel-Ittah, Danny Ben-Zvi, Eyal D. Schejter, Naama Barkai. Morphogen gradients are used to pattern a field of cells according to variations in the concentration of a signaling molecule. Typically, the morphogen emanates from a confined group of cells. Dur....
Thursday, January 24, 2013
[Report] Tunable Signal Processing Through Modular Control of Transcription Factor Translocation
[Report] Tunable Signal Processing Through Modular Control of Transcription Factor Translocation: A yeast protein transforms stress signals into distinct dynamic responses according to the timing and strength of inputs.
Authors: Nan Hao, Bogdan A. Budnik, Jeremy Gunawardena, Erin K. O'Shea
Authors: Nan Hao, Bogdan A. Budnik, Jeremy Gunawardena, Erin K. O'Shea
Insights into bilaterian evolution from three spiralian genomes
Insights into bilaterian evolution from three spiralian genomes:
Insights into bilaterian evolution from three spiralian genomes
Nature 493, 7433 (2013). doi:10.1038/nature11696
Authors: Oleg Simakov, Ferdinand Marletaz, Sung-Jin Cho, Eric Edsinger-Gonzales, Paul Havlak, Uffe Hellsten, Dian-Han Kuo, Tomas Larsson, Jie Lv, Detlev Arendt, Robert Savage, Kazutoyo Osoegawa, Pieter de Jong, Jane Grimwood, Jarrod A. Chapman, Harris Shapiro, Andrea Aerts, Robert P. Otillar, Astrid Y. Terry, Jeffrey L. Boore, Igor V. Grigoriev, David R. Lindberg, Elaine C. Seaver, David A. Weisblat, Nicholas H. Putnam & Daniel S. Rokhsar
Current genomic perspectives on animal diversity neglect two prominent phyla, the molluscs and annelids, that together account for nearly one-third of known marine species and are important both ecologically and as experimental systems in classical embryology. Here we describe the draft genomes of the owl limpet (Lottia gigantea), a marine polychaete (Capitella teleta) and a freshwater leech (Helobdella robusta), and compare them with other animal genomes to investigate the origin and diversification of bilaterians from a genomic perspective. We find that the genome organization, gene structure and functional content of these species are more similar to those of some invertebrate deuterostome genomes (for example, amphioxus and sea urchin) than those of other protostomes that have been sequenced to date (flies, nematodes and flatworms). The conservation of these genomic features enables us to expand the inventory of genes present in the last common bilaterian ancestor, establish the tripartite diversification of bilaterians using multiple genomic characteristics and identify ancient conserved long- and short-range genetic linkages across metazoans. Superimposed on this broadly conserved pan-bilaterian background we find examples of lineage-specific genome evolution, including varying rates of rearrangement, intron gain and loss, expansions and contractions of gene families, and the evolution of clade-specific genes that produce the unique content of each genome.

Insights into bilaterian evolution from three spiralian genomes
Nature 493, 7433 (2013). doi:10.1038/nature11696
Authors: Oleg Simakov, Ferdinand Marletaz, Sung-Jin Cho, Eric Edsinger-Gonzales, Paul Havlak, Uffe Hellsten, Dian-Han Kuo, Tomas Larsson, Jie Lv, Detlev Arendt, Robert Savage, Kazutoyo Osoegawa, Pieter de Jong, Jane Grimwood, Jarrod A. Chapman, Harris Shapiro, Andrea Aerts, Robert P. Otillar, Astrid Y. Terry, Jeffrey L. Boore, Igor V. Grigoriev, David R. Lindberg, Elaine C. Seaver, David A. Weisblat, Nicholas H. Putnam & Daniel S. Rokhsar
Current genomic perspectives on animal diversity neglect two prominent phyla, the molluscs and annelids, that together account for nearly one-third of known marine species and are important both ecologically and as experimental systems in classical embryology. Here we describe the draft genomes of the owl limpet (Lottia gigantea), a marine polychaete (Capitella teleta) and a freshwater leech (Helobdella robusta), and compare them with other animal genomes to investigate the origin and diversification of bilaterians from a genomic perspective. We find that the genome organization, gene structure and functional content of these species are more similar to those of some invertebrate deuterostome genomes (for example, amphioxus and sea urchin) than those of other protostomes that have been sequenced to date (flies, nematodes and flatworms). The conservation of these genomic features enables us to expand the inventory of genes present in the last common bilaterian ancestor, establish the tripartite diversification of bilaterians using multiple genomic characteristics and identify ancient conserved long- and short-range genetic linkages across metazoans. Superimposed on this broadly conserved pan-bilaterian background we find examples of lineage-specific genome evolution, including varying rates of rearrangement, intron gain and loss, expansions and contractions of gene families, and the evolution of clade-specific genes that produce the unique content of each genome.
Saturday, January 19, 2013
Promoter cross-talk via a shared enhancer explains paternally biased expression of Nctc1 at the Igf2/H19/Nctc1 imprinted locus
Promoter cross-talk via a shared enhancer explains paternally biased expression of Nctc1 at the Igf2/H19/Nctc1 imprinted locus:
Developmentally regulated transcription often depends on physical interactions between distal enhancers and their cognate promoters. Recent genomic analyses suggest that promoter–promoter interactions might play a similarly critical role in organizing the genome and establishing cell-type-specific gene expression. The Igf2/H19 locus has been a valuable model for clarifying the role of long-range interactions between cis-regulatory elements. Imprinted expression of the linked, reciprocally imprinted genes is explained by parent-of-origin-specific chromosomal loop structures between the paternal Igf2 or maternal H19 promoters and their shared tissue-specific enhancer elements. Here, we further analyze these loop structures for their composition and their impact on expression of the linked long non-coding RNA, Nctc1. We show that Nctc1 is co-regulated with Igf2 and H19 and physically interacts with the shared muscle enhancer. In fact, all three co-regulated genes have the potential to interact not only with the shared enhancer but also with each other via their enhancer interactions. Furthermore, developmental and genetic analyses indicate functional significance for these promoter–promoter interactions. Altogether, we present a novel mechanism to explain developmental specific imprinting of Nctc1 and provide new information about enhancer mechanisms and about the role of chromatin domains in establishing gene expression patterns.
Developmentally regulated transcription often depends on physical interactions between distal enhancers and their cognate promoters. Recent genomic analyses suggest that promoter–promoter interactions might play a similarly critical role in organizing the genome and establishing cell-type-specific gene expression. The Igf2/H19 locus has been a valuable model for clarifying the role of long-range interactions between cis-regulatory elements. Imprinted expression of the linked, reciprocally imprinted genes is explained by parent-of-origin-specific chromosomal loop structures between the paternal Igf2 or maternal H19 promoters and their shared tissue-specific enhancer elements. Here, we further analyze these loop structures for their composition and their impact on expression of the linked long non-coding RNA, Nctc1. We show that Nctc1 is co-regulated with Igf2 and H19 and physically interacts with the shared muscle enhancer. In fact, all three co-regulated genes have the potential to interact not only with the shared enhancer but also with each other via their enhancer interactions. Furthermore, developmental and genetic analyses indicate functional significance for these promoter–promoter interactions. Altogether, we present a novel mechanism to explain developmental specific imprinting of Nctc1 and provide new information about enhancer mechanisms and about the role of chromatin domains in establishing gene expression patterns.
Integrative annotation of chromatin elements from ENCODE data
Integrative annotation of chromatin elements from ENCODE data:
The ENCODE Project has generated a wealth of experimental information mapping diverse chromatin properties in several human cell lines. Although each such data track is independently informative toward the annotation of regulatory elements, their interrelations contain much richer information for the systematic annotation of regulatory elements. To uncover these interrelations and to generate an interpretable summary of the massive datasets of the ENCODE Project, we apply unsupervised learning methodologies, converting dozens of chromatin datasets into discrete annotation maps of regulatory regions and other chromatin elements across the human genome. These methods rediscover and summarize diverse aspects of chromatin architecture, elucidate the interplay between chromatin activity and RNA transcription, and reveal that a large proportion of the genome lies in a quiescent state, even across multiple cell types. The resulting annotation of non-coding regulatory elements correlate strongly with mammalian evolutionary constraint, and provide an unbiased approach for evaluating metrics of evolutionary constraint in human. Lastly, we use the regulatory annotations to revisit previously uncharacterized disease-associated loci, resulting in focused, testable hypotheses through the lens of the chromatin landscape.
The ENCODE Project has generated a wealth of experimental information mapping diverse chromatin properties in several human cell lines. Although each such data track is independently informative toward the annotation of regulatory elements, their interrelations contain much richer information for the systematic annotation of regulatory elements. To uncover these interrelations and to generate an interpretable summary of the massive datasets of the ENCODE Project, we apply unsupervised learning methodologies, converting dozens of chromatin datasets into discrete annotation maps of regulatory regions and other chromatin elements across the human genome. These methods rediscover and summarize diverse aspects of chromatin architecture, elucidate the interplay between chromatin activity and RNA transcription, and reveal that a large proportion of the genome lies in a quiescent state, even across multiple cell types. The resulting annotation of non-coding regulatory elements correlate strongly with mammalian evolutionary constraint, and provide an unbiased approach for evaluating metrics of evolutionary constraint in human. Lastly, we use the regulatory annotations to revisit previously uncharacterized disease-associated loci, resulting in focused, testable hypotheses through the lens of the chromatin landscape.
Conserved boundary elements from the Hox complex of mosquito, Anopheles gambiae
Conserved boundary elements from the Hox complex of mosquito, Anopheles gambiae:
The conservation of hox genes as well as their genomic organization across the phyla suggests that this system of anterior–posterior axis formation arose early during evolution and has come under strong selection pressure. Studies in the split Hox cluster of Drosophila have shown that proper expression of hox genes is dependent on chromatin domain boundaries that prevent inappropriate interactions among different types of cis-regulatory elements. To investigate whether boundary function and their role in regulation of hox genes is conserved in insects with intact Hox clusters, we used an algorithm to locate potential boundary elements in the Hox complex of mosquito, Anopheles gambiae. Several potential boundary elements were identified that could be tested for their functional conservation. Comparative analysis revealed that like Drosophila, the bithorax region in A. gambiae contains an extensive array of boundaries and enhancers organized into domains. We analysed a subset of candidate boundary elements and show that they function as enhancer blockers in Drosophila. The functional conservation of boundary elements from mosquito in fly suggests that regulation of hox genes involving chromatin domain boundaries is an evolutionary conserved mechanism and points to an important role of such elements in key developmentally regulated loci.
The conservation of hox genes as well as their genomic organization across the phyla suggests that this system of anterior–posterior axis formation arose early during evolution and has come under strong selection pressure. Studies in the split Hox cluster of Drosophila have shown that proper expression of hox genes is dependent on chromatin domain boundaries that prevent inappropriate interactions among different types of cis-regulatory elements. To investigate whether boundary function and their role in regulation of hox genes is conserved in insects with intact Hox clusters, we used an algorithm to locate potential boundary elements in the Hox complex of mosquito, Anopheles gambiae. Several potential boundary elements were identified that could be tested for their functional conservation. Comparative analysis revealed that like Drosophila, the bithorax region in A. gambiae contains an extensive array of boundaries and enhancers organized into domains. We analysed a subset of candidate boundary elements and show that they function as enhancer blockers in Drosophila. The functional conservation of boundary elements from mosquito in fly suggests that regulation of hox genes involving chromatin domain boundaries is an evolutionary conserved mechanism and points to an important role of such elements in key developmentally regulated loci.
Friday, January 18, 2013
DNA-Binding Specificities of Human Transcription Factors
DNA-Binding Specificities of Human Transcription Factors: Arttu Jolma, Jian Yan, Thomas Whitington, Jarkko Toivonen, Kazuhiro R. Nitta, Pasi Rastas, Ekaterina Morgunova, Martin Enge, Mikko Taipale, Gonghong Wei, Kimmo Palin, Juan M. Vaquerizas, Renaud Vincentelli, Nicholas M. Luscombe, Timothy R. Hughes, Patrick Lemaire, Esko Ukkonen, Teemu Kivioja, Jussi Taipale. Although the proteins that read the gene regulatory code, transcription factors (TFs), have been largely identified, it is not well known which sequences TFs can recognize. We have analyzed the se....
Latent Enhancers Activated by Stimulation in Differentiated Cells
Latent Enhancers Activated by Stimulation in Differentiated Cells: Renato Ostuni, Viviana Piccolo, Iros Barozzi, Sara Polletti, Alberto Termanini, Silvia Bonifacio, Alessia Curina, Elena Prosperini, Serena Ghisletti, Gioacchino Natoli. According to current models, once the cell has reached terminal differentiation, the enhancer repertoire is completely established and maintained by cooperatively acting lineage-specific transcrip....
System-wide Rewiring Underlies Behavioral Differences in Predatory and Bacterial-Feeding Nematodes
System-wide Rewiring Underlies Behavioral Differences in Predatory and Bacterial-Feeding Nematodes: Daniel J. Bumbarger, Metta Riebesell, Christian Rödelsperger, Ralf J. Sommer. The relationship between neural circuit function and patterns of synaptic connectivity is poorly understood, in part due to a lack of comparative data for larger complete systems. We compare syste....
Developmentally Regulated Subnuclear Genome Reorganization Restricts Neural Progenitor Competence in Drosophila
Developmentally Regulated Subnuclear Genome Reorganization Restricts Neural Progenitor Competence in Drosophila: Minoree Kohwi, Joshua R. Lupton, Sen-Lin Lai, Michael R. Miller, Chris Q. Doe. Stem and/or progenitor cells often generate distinct cell types in a stereotyped birth order and over time lose competence to specify earlier-born fates by unknown mechanisms. In Drosophila....
Genome-wide Chromatin State Transitions Associated with Developmental and Environmental Cues
Genome-wide Chromatin State Transitions Associated with Developmental and Environmental Cues: Jiang Zhu, Mazhar Adli, James Y. Zou, Griet Verstappen, Michael Coyne, Xiaolan Zhang, Timothy Durham, Mohammad Miri, Vikram Deshpande, Philip L. De Jager, David A. Bennett, Joseph A. Houmard, Deborah M. Muoio, Tamer T. Onder, Ray Camahort, Chad A. Cowan, Alexander Meissner, Charles B. Epstein, Noam Shoresh, Bradley E. Bernstein.
Differences in chromatin organization are key to the multiplicity of cell states that arise from a single genetic background, yet the landscapes of in vivo tissues remain largely uncharted. Here, ....
Differences in chromatin organization are key to the multiplicity of cell states that arise from a single genetic background, yet the landscapes of in vivo tissues remain largely uncharted. Here, ....
Wednesday, January 16, 2013
Discrete genetic modules are responsible for complex burrow evolution in Peromyscus mice
Discrete genetic modules are responsible for complex burrow evolution in Peromyscus mice:
Discrete genetic modules are responsible for complex burrow evolution in Peromyscus mice
Nature 493, 7432 (2013). doi:10.1038/nature11816
Authors: Jesse N. Weber, Brant K. Peterson & Hopi E. Hoekstra
Relative to morphological traits, we know little about how genetics influence the evolution of complex behavioural differences in nature. It is unclear how the environment influences natural variation in heritable behaviour, and whether complex behavioural differences evolve through few genetic changes, each affecting many aspects of behaviour, or through the accumulation of several genetic changes that, when combined, give rise to behavioural complexity. Here we show that in nature, oldfield mice (Peromyscus polionotus) build complex burrows with long entrance and escape tunnels, and that burrow length is consistent across populations, although burrow depth varies with soil composition. This burrow architecture is in contrast with the small, simple burrows of its sister species, deer mice (P. maniculatus). When investigated under laboratory conditions, both species recapitulate their natural burrowing behaviour. Genetic crosses between the two species reveal that the derived burrows of oldfield mice are dominant and evolved through the addition of multiple genetic changes. In burrows built by first-generation backcross mice, entrance-tunnel length and the presence of an escape tunnel can be uncoupled, suggesting that these traits are modular. Quantitative trait locus analysis also indicates that tunnel length segregates as a complex trait, affected by at least three independent genetic regions, whereas the presence of an escape tunnel is associated with only a single locus. Together, these results suggest that complex behaviours—in this case, a classic ‘extended phenotype’—can evolve through multiple genetic changes each affecting distinct behaviour modules.

Discrete genetic modules are responsible for complex burrow evolution in Peromyscus mice
Nature 493, 7432 (2013). doi:10.1038/nature11816
Authors: Jesse N. Weber, Brant K. Peterson & Hopi E. Hoekstra
Relative to morphological traits, we know little about how genetics influence the evolution of complex behavioural differences in nature. It is unclear how the environment influences natural variation in heritable behaviour, and whether complex behavioural differences evolve through few genetic changes, each affecting many aspects of behaviour, or through the accumulation of several genetic changes that, when combined, give rise to behavioural complexity. Here we show that in nature, oldfield mice (Peromyscus polionotus) build complex burrows with long entrance and escape tunnels, and that burrow length is consistent across populations, although burrow depth varies with soil composition. This burrow architecture is in contrast with the small, simple burrows of its sister species, deer mice (P. maniculatus). When investigated under laboratory conditions, both species recapitulate their natural burrowing behaviour. Genetic crosses between the two species reveal that the derived burrows of oldfield mice are dominant and evolved through the addition of multiple genetic changes. In burrows built by first-generation backcross mice, entrance-tunnel length and the presence of an escape tunnel can be uncoupled, suggesting that these traits are modular. Quantitative trait locus analysis also indicates that tunnel length segregates as a complex trait, affected by at least three independent genetic regions, whereas the presence of an escape tunnel is associated with only a single locus. Together, these results suggest that complex behaviours—in this case, a classic ‘extended phenotype’—can evolve through multiple genetic changes each affecting distinct behaviour modules.
Tuesday, January 15, 2013
Drosophila Left/Right Asymmetry Establishment Is Controlled by the Hox Gene Abdominal-B
Drosophila Left/Right Asymmetry Establishment Is Controlled by the Hox Gene Abdominal-B: Jean-Baptiste Coutelis, Charles Géminard, Pauline Spéder, Magali Suzanne, Astrid Gerlinde Petzoldt, Stéphane Noselli. In Drosophila, left/right (LR) asymmetry is apparent in the directional looping of the gut and male genitalia. The dextral orientation of the organs depends on the activity of a single gene....
Robustness and Epistasis in the C. elegans Vulval Signaling Network Revealed by Pathway Dosage Modulation
Robustness and Epistasis in the C. elegans Vulval Signaling Network Revealed by Pathway Dosage Modulation: Michalis Barkoulas, Jeroen S. van Zon, Josselin Milloz, Alexander van Oudenaarden, Marie-Anne Félix. Biological systems may perform reproducibly to generate invariant outcomes, despite external or internal noise. One example is the C. elegans vulva, in which the final cell fate pattern is ....
Thursday, January 10, 2013
Genome-Wide Screens for In Vivo Tinman Binding Sites Identify Cardiac Enhancers with Diverse Functional Architectures
Genome-Wide Screens for In Vivo Tinman Binding Sites Identify Cardiac Enhancers with Diverse Functional Architectures:
by Hong Jin, Robert Stojnic, Boris Adryan, Anil Ozdemir, Angelike Stathopoulos, Manfred Frasch
The NK homeodomain factor Tinman is a crucial regulator of early mesoderm patterning and, together with the GATA factor Pannier and the Dorsocross T-box factors, serves as one of the key cardiogenic factors during specification and differentiation of heart cells. Although the basic framework of regulatory interactions driving heart development has been worked out, only about a dozen genes involved in heart development have been designated as direct Tinman target genes to date, and detailed information about the functional architectures of their cardiac enhancers is lacking. We have used immunoprecipitation of chromatin (ChIP) from embryos at two different stages of early cardiogenesis to obtain a global overview of the sequences bound by Tinman in vivo and their linked genes. Our data from the analysis of ∼50 sequences with high Tinman occupancy show that the majority of such sequences act as enhancers in various mesodermal tissues in which Tinman is active. All of the dorsal mesodermal and cardiac enhancers, but not some of the others, require tinman function. The cardiac enhancers feature diverse arrangements of binding motifs for Tinman, Pannier, and Dorsocross. By employing these cardiac and non-cardiac enhancers in machine learning approaches, we identify a novel motif, termed CEE, as a classifier for cardiac enhancers. In vivo assays for the requirement of the binding motifs of Tinman, Pannier, and Dorsocross, as well as the CEE motifs in a set of cardiac enhancers, show that the Tinman sites are essential in all but one of the tested enhancers; although on occasion they can be functionally redundant with Dorsocross sites. The enhancers differ widely with respect to their requirement for Pannier, Dorsocross, and CEE sites, which we ascribe to their different position in the regulatory circuitry, their distinct temporal and spatial activities during cardiogenesis, and functional redundancies among different factor binding sites.
by Hong Jin, Robert Stojnic, Boris Adryan, Anil Ozdemir, Angelike Stathopoulos, Manfred Frasch
The NK homeodomain factor Tinman is a crucial regulator of early mesoderm patterning and, together with the GATA factor Pannier and the Dorsocross T-box factors, serves as one of the key cardiogenic factors during specification and differentiation of heart cells. Although the basic framework of regulatory interactions driving heart development has been worked out, only about a dozen genes involved in heart development have been designated as direct Tinman target genes to date, and detailed information about the functional architectures of their cardiac enhancers is lacking. We have used immunoprecipitation of chromatin (ChIP) from embryos at two different stages of early cardiogenesis to obtain a global overview of the sequences bound by Tinman in vivo and their linked genes. Our data from the analysis of ∼50 sequences with high Tinman occupancy show that the majority of such sequences act as enhancers in various mesodermal tissues in which Tinman is active. All of the dorsal mesodermal and cardiac enhancers, but not some of the others, require tinman function. The cardiac enhancers feature diverse arrangements of binding motifs for Tinman, Pannier, and Dorsocross. By employing these cardiac and non-cardiac enhancers in machine learning approaches, we identify a novel motif, termed CEE, as a classifier for cardiac enhancers. In vivo assays for the requirement of the binding motifs of Tinman, Pannier, and Dorsocross, as well as the CEE motifs in a set of cardiac enhancers, show that the Tinman sites are essential in all but one of the tested enhancers; although on occasion they can be functionally redundant with Dorsocross sites. The enhancers differ widely with respect to their requirement for Pannier, Dorsocross, and CEE sites, which we ascribe to their different position in the regulatory circuitry, their distinct temporal and spatial activities during cardiogenesis, and functional redundancies among different factor binding sites.
Wednesday, January 9, 2013
Evolutionary Biology for the 21st Century
Evolutionary Biology for the 21st Century:
by Jonathan B. Losos, Stevan J. Arnold, Gill Bejerano, E. D. Brodie, David Hibbett, Hopi E. Hoekstra, David P. Mindell, Antónia Monteiro, Craig Moritz, H. Allen Orr, Dmitri A. Petrov, Susanne S. Renner, Robert E. Ricklefs, Pamela S. Soltis, Thomas L. Turner

by Jonathan B. Losos, Stevan J. Arnold, Gill Bejerano, E. D. Brodie, David Hibbett, Hopi E. Hoekstra, David P. Mindell, Antónia Monteiro, Craig Moritz, H. Allen Orr, Dmitri A. Petrov, Susanne S. Renner, Robert E. Ricklefs, Pamela S. Soltis, Thomas L. Turner
Sunday, January 6, 2013
Transcriptional repressors: multifaceted regulators of gene expression [REVIEW]
Transcriptional repressors: multifaceted regulators of gene expression [REVIEW]: Nicola Reynolds, Aoife O'Shaughnessy, and Brian Hendrich
Through decades of research it has been established that some chromatin-modifying proteins can repress transcription, and thus are generally termed ‘repressors’. Although classic repressors undoubtedly silence transcription, genome-wide studies have shown that many repressors are associated with actively transcribed loci and that this is a widespread phenomenon. Here, we review the evidence for the presence of repressors at actively transcribed regions and assess what roles they might be playing. We propose that the modulation of expression levels by chromatin-modifying, co-repressor complexes provides transcriptional fine-tuning that drives development.
Through decades of research it has been established that some chromatin-modifying proteins can repress transcription, and thus are generally termed ‘repressors’. Although classic repressors undoubtedly silence transcription, genome-wide studies have shown that many repressors are associated with actively transcribed loci and that this is a widespread phenomenon. Here, we review the evidence for the presence of repressors at actively transcribed regions and assess what roles they might be playing. We propose that the modulation of expression levels by chromatin-modifying, co-repressor complexes provides transcriptional fine-tuning that drives development.
Establishing and maintaining gene expression patterns: insights from sensory receptor patterning [PRIMER]
Establishing and maintaining gene expression patterns: insights from sensory receptor patterning [PRIMER]: Jens Rister, Claude Desplan, and Daniel Vasiliauskas
In visual and olfactory sensory systems with high discriminatory power, each sensory neuron typically expresses one, or very few, sensory receptor genes, excluding all others. Recent studies have provided insights into the mechanisms that generate and maintain sensory receptor expression patterns. Here, we review how this is achieved in the fly retina and compare it with the mechanisms controlling sensory receptor expression patterns in the mouse retina and in the mouse and fly olfactory systems.
In visual and olfactory sensory systems with high discriminatory power, each sensory neuron typically expresses one, or very few, sensory receptor genes, excluding all others. Recent studies have provided insights into the mechanisms that generate and maintain sensory receptor expression patterns. Here, we review how this is achieved in the fly retina and compare it with the mechanisms controlling sensory receptor expression patterns in the mouse retina and in the mouse and fly olfactory systems.
Friday, January 4, 2013
Number of Nuclear Divisions in the Drosophila Blastoderm Controlled by Onset of Zygotic Transcription
Number of Nuclear Divisions in the Drosophila Blastoderm Controlled by Onset of Zygotic Transcription: Hung-wei Sung, Saskia Spangenberg, Nina Vogt, Jörg Großhans. The cell number of the early Drosophila embryo is determined by exactly 13 rounds of synchronous nuclear divisions, allowing cellularization and formation of the embryonic epithelium [1]. T....
Thursday, January 3, 2013
The Underlying Molecular and Network Level Mechanisms in the Evolution of Robustness in Gene Regulatory Networks
The Underlying Molecular and Network Level Mechanisms in the Evolution of Robustness in Gene Regulatory Networks:
by Mario Pujato, Thomas MacCarthy, Andras Fiser, Aviv Bergman
Gene regulatory networks show robustness to perturbations. Previous works identified robustness as an emergent property of gene network evolution but the underlying molecular mechanisms are poorly understood. We used a multi-tier modeling approach that integrates molecular sequence and structure information with network architecture and population dynamics. Structural models of transcription factor-DNA complexes are used to estimate relative binding specificities. In this model, mutations in the DNA cause changes on two levels: (a) at the sequence level in individual binding sites (modulating binding specificity), and (b) at the network level (creating and destroying binding sites). We used this model to dissect the underlying mechanisms responsible for the evolution of robustness in gene regulatory networks. Results suggest that in sparse architectures (represented by short promoters), a mixture of local-sequence and network-architecture level changes are exploited. At the local-sequence level, robustness evolves by decreasing the probabilities of both the destruction of existent and generation of new binding sites. Meanwhile, in highly interconnected architectures (represented by long promoters), robustness evolves almost entirely via network level changes, deleting and creating binding sites that modify the network architecture.
by Mario Pujato, Thomas MacCarthy, Andras Fiser, Aviv Bergman
Gene regulatory networks show robustness to perturbations. Previous works identified robustness as an emergent property of gene network evolution but the underlying molecular mechanisms are poorly understood. We used a multi-tier modeling approach that integrates molecular sequence and structure information with network architecture and population dynamics. Structural models of transcription factor-DNA complexes are used to estimate relative binding specificities. In this model, mutations in the DNA cause changes on two levels: (a) at the sequence level in individual binding sites (modulating binding specificity), and (b) at the network level (creating and destroying binding sites). We used this model to dissect the underlying mechanisms responsible for the evolution of robustness in gene regulatory networks. Results suggest that in sparse architectures (represented by short promoters), a mixture of local-sequence and network-architecture level changes are exploited. At the local-sequence level, robustness evolves by decreasing the probabilities of both the destruction of existent and generation of new binding sites. Meanwhile, in highly interconnected architectures (represented by long promoters), robustness evolves almost entirely via network level changes, deleting and creating binding sites that modify the network architecture.
Wednesday, January 2, 2013
A second-generation assembly of the Drosophila simulans genome provides new insights into patterns of lineage-specific divergence [RESEARCH]
A second-generation assembly of the Drosophila simulans genome provides new insights into patterns of lineage-specific divergence [RESEARCH]:
We create a new assembly of the Drosophila simulans genome using 142 million paired short-read sequences and previously published data for strain w501. Our assembly represents a higher-quality genomic sequence with greater coverage, fewer misassemblies, and, by several indexes, fewer sequence errors. Evolutionary analysis of this genome reference sequence reveals interesting patterns of lineage-specific divergence that are different from those previously reported. Specifically, we find that Drosophila melanogaster evolves faster than D. simulans at all annotated classes of sites, including putatively neutrally evolving sites found in minimal introns. While this may be partly explained by a higher mutation rate in D. melanogaster, we also find significant heterogeneity in rates of evolution across classes of sites, consistent with historical differences in the effective population size for the two species. Also contrary to previous findings, we find that the X chromosome is evolving significantly faster than autosomes for nonsynonymous and most noncoding DNA sites and significantly slower for synonymous sites. The absence of a X/A difference for putatively neutral sites and the robustness of the pattern to Gene Ontology and sex-biased expression suggest that partly recessive beneficial mutations may comprise a substantial fraction of noncoding DNA divergence observed between species. Our results have more general implications for the interpretation of evolutionary analyses of genomes of different quality.
We create a new assembly of the Drosophila simulans genome using 142 million paired short-read sequences and previously published data for strain w501. Our assembly represents a higher-quality genomic sequence with greater coverage, fewer misassemblies, and, by several indexes, fewer sequence errors. Evolutionary analysis of this genome reference sequence reveals interesting patterns of lineage-specific divergence that are different from those previously reported. Specifically, we find that Drosophila melanogaster evolves faster than D. simulans at all annotated classes of sites, including putatively neutrally evolving sites found in minimal introns. While this may be partly explained by a higher mutation rate in D. melanogaster, we also find significant heterogeneity in rates of evolution across classes of sites, consistent with historical differences in the effective population size for the two species. Also contrary to previous findings, we find that the X chromosome is evolving significantly faster than autosomes for nonsynonymous and most noncoding DNA sites and significantly slower for synonymous sites. The absence of a X/A difference for putatively neutral sites and the robustness of the pattern to Gene Ontology and sex-biased expression suggest that partly recessive beneficial mutations may comprise a substantial fraction of noncoding DNA divergence observed between species. Our results have more general implications for the interpretation of evolutionary analyses of genomes of different quality.
GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility [RESEARCH]
GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility [RESEARCH]:
Estrogen receptor (ESR1) drives growth in the majority of human breast cancers by binding to regulatory elements and inducing transcription events that promote tumor growth. Differences in enhancer occupancy by ESR1 contribute to the diverse expression profiles and clinical outcome observed in breast cancer patients. GATA3 is an ESR1-cooperating transcription factor mutated in breast tumors; however, its genomic properties are not fully defined. In order to investigate the composition of enhancers involved in estrogen-induced transcription and the potential role of GATA3, we performed extensive ChIP-sequencing in unstimulated breast cancer cells and following estrogen treatment. We find that GATA3 is pivotal in mediating enhancer accessibility at regulatory regions involved in ESR1-mediated transcription. GATA3 silencing resulted in a global redistribution of cofactors and active histone marks prior to estrogen stimulation. These global genomic changes altered the ESR1-binding profile that subsequently occurred following estrogen, with events exhibiting both loss and gain in binding affinity, implying a GATA3-mediated redistribution of ESR1 binding. The GATA3-mediated redistributed ESR1 profile correlated with changes in gene expression, suggestive of its functionality. Chromatin loops at the TFF locus involving ESR1-bound enhancers occurred independently of ESR1 when GATA3 was silenced, indicating that GATA3, when present on the chromatin, may serve as a licensing factor for estrogen–ESR1-mediated interactions between cis-regulatory elements. Together, these experiments suggest that GATA3 directly impacts ESR1 enhancer accessibility, and may potentially explain the contribution of mutant-GATA3 in the heterogeneity of ESR1+ breast cancer.
Estrogen receptor (ESR1) drives growth in the majority of human breast cancers by binding to regulatory elements and inducing transcription events that promote tumor growth. Differences in enhancer occupancy by ESR1 contribute to the diverse expression profiles and clinical outcome observed in breast cancer patients. GATA3 is an ESR1-cooperating transcription factor mutated in breast tumors; however, its genomic properties are not fully defined. In order to investigate the composition of enhancers involved in estrogen-induced transcription and the potential role of GATA3, we performed extensive ChIP-sequencing in unstimulated breast cancer cells and following estrogen treatment. We find that GATA3 is pivotal in mediating enhancer accessibility at regulatory regions involved in ESR1-mediated transcription. GATA3 silencing resulted in a global redistribution of cofactors and active histone marks prior to estrogen stimulation. These global genomic changes altered the ESR1-binding profile that subsequently occurred following estrogen, with events exhibiting both loss and gain in binding affinity, implying a GATA3-mediated redistribution of ESR1 binding. The GATA3-mediated redistributed ESR1 profile correlated with changes in gene expression, suggestive of its functionality. Chromatin loops at the TFF locus involving ESR1-bound enhancers occurred independently of ESR1 when GATA3 was silenced, indicating that GATA3, when present on the chromatin, may serve as a licensing factor for estrogen–ESR1-mediated interactions between cis-regulatory elements. Together, these experiments suggest that GATA3 directly impacts ESR1 enhancer accessibility, and may potentially explain the contribution of mutant-GATA3 in the heterogeneity of ESR1+ breast cancer.
Subscribe to:
Posts (Atom)