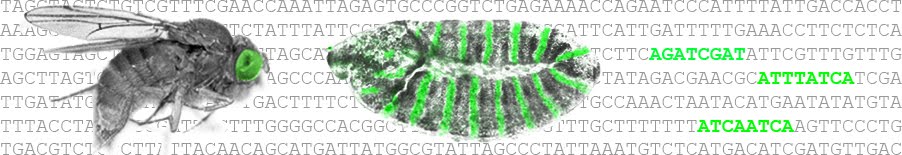
Genes responsible for quantitative variation have been identified for a diverse range of phenotypes. However, much remains to be learned about the distribution of causative genetic variation within a locus. In this study, we investigated a locus that contributes to natural variation in abdominal pigmentation in Drosophila melanogaster. We found that the large phenotypic effect of this locus results from the cumulative action of many small-effect polymorphisms that are concentrated in three distinct functional regions: a promoter, a tissue-specific enhancer, and a Polycomb response element (a region involved in chromatin remodeling). The same regions influence the adult phenotype and transcript abundance, indicating that the causative sequence variants act by modulating transcription. Interestingly, these polymorphisms cluster near, but not within, the functionally validated regulatory regions, suggesting that DNA sequences surrounding core functional elements may play a key role in quantitative variation.
Monday, January 24, 2011
Composite Effects of Polymorphisms near Multiple Regulatory Elements Create a Major-Effect QTL
Friday, January 21, 2011
Global Analysis of the Impact of Environmental Perturbation on cis-Regulation of Gene Expression
Population variation in normal gene expression has been convincingly shown to be under strong genetic control where the main genetic variants are located within close proximity to the gene itself (so called cis-acting). However, the extent to which controlled, environmental stimuli influences cis-regulation of gene expression is unclear. Here, we combine different functional genomic approaches and examine the role of common genetic variants on induced gene expression in a population panel of primary human cells derived from ~100 unrelated donors treated under multiple conditions. Using these approaches, we find that the interaction between cellular environment and cis-variants are relatively rare, with only a small proportion of the identified genetic variants being specific to treatment. However, although treatment-specific genetic regulation of gene expression seems to be infrequent, we prove its existence by thorough validation of treatment-specific effects of the glucocorticoid-specific regulation of TNC expression. Taken together, these findings indicate that the regulatory landscape within a cell is very stable but, by combining functional genomic tools gene-environmental interactions of clinical importance, can be detected and possibly used as biomarkers in future pharmacogenomic studies.
Segregating Variation in the Polycomb Group Gene cramped Alters the Effect of Temperature on Multiple Traits
The fruitfly Drosophila melanogaster was initially endemic to sub-Saharan Africa and started to colonize Europe and Asia around 10,000 years ago. Northern populations have adapted to these colder environments and differ from Sub-Saharan populations for temperature sensitive traits. Here we analyze cramped (crm), a gene previously identified as a putative target of adaptive selection during the colonization of northern latitudes. crm is involved in the regulation of chromosome structure, a process known to be strongly modulated by temperature. We show that crm is limiting for distinct processes at different temperatures and that crm natural variation modulates temperature sensitive phenotypes. Our results suggest that environmental heterogeneity maintains functional variation in environment sensitive gene networks and might facilitate evolution.
Tuesday, January 11, 2011
cis-regulatory mutations are a genetic cause of human limb malformations
Abstract
The underlying mutations that cause human limb malformations are often difficult to determine, particularly for limb malformations that occur as isolated traits. Evidence from a variety of studies shows that cis-regulatory mutations, specifically in enhancers, can lead to some of these isolated limb malformations. Here, we provide a review of human limb malformations that have been shown to be caused by enhancer mutations and propose that cis-regulatory mutations will continue to be identified as the cause of additional human malformations as our understanding of regulatory sequences improves. Developmental Dynamics, 2011. © 2011 Wiley-Liss, Inc.
Genomic Differentiation Between Temperate and Tropical Australian Populations of Drosophila melanogaster [Population and evolutionary genetics]
Determining the genetic basis of environmental adaptation is a central problem of evolutionary biology. This issue has been fruitfully addressed by examining genetic differentiation between populations that are recently separated and/or experience high rates of gene flow. A good example of this approach is the decades-long investigation of selection acting along latitudinal clines in Drosophila melanogaster. Here we use next-generation genome sequencing to reexamine the well-studied Australian D. melanogaster cline. We find evidence for extensive differentiation between temperate and tropical populations, with regulatory regions and unannotated regions showing particularly high levels of differentiation. Although the physical genomic scale of geographic differentiation is small—on the order of gene sized—we observed several larger highly differentiated regions. The region spanned by the cosmopolitan inversion polymorphism In(3R)P shows higher levels of differentiation, consistent with the major difference in allele frequencies of Standard and In(3R)P karyotypes in temperate vs. tropical Australian populations. Our analysis reveals evidence for spatially varying selection on a number of key biological processes, suggesting fundamental biological differences between flies from these two geographic regions.
"Coordinated Genome-Wide Modifications within Proximal Promoter Cis-regulatory Elements during Vertebrate Evolution
There often exists a 'one-to-many' relationship between a transcription factor and a multitude of binding sites throughout the genome. It is commonly assumed that transcription factor binding motifs remain largely static over the course of evolution because changes in binding specificity can alter the interactions with potentially hundreds of sites across the genome. Focusing on regulatory motifs overrepresented at specific locations within or near the promoter, we find that a surprisingly large number of cis-regulatory elements have been subject to coordinated genome-wide modifications during vertebrate evolution, such that the motif frequency changes on a single branch of vertebrate phylogeny. This was found to be the case even between closely related mammal species, with nearly a third of all location-specific consensus motifs exhibiting significant modifications within the human or mouse lineage since their divergence. Many of these modifications are likely to be compensatory changes throughout the genome following changes in protein factor binding affinities, whereas others may be due to changes in mutation rates or effective population size. The likelihood that this happened many times during vertebrate evolution highlights the need to examine additional taxa and to understand the evolutionary and molecular mechanisms underlying the evolution of protein–DNA interactions.
"Wednesday, January 5, 2011
Identification of cis- and trans-regulatory variation modulating microRNA expression levels in human fibroblasts [RESEARCH]
MicroRNAs (miRNAs) are regulatory noncoding RNAs that affect the production of a significant fraction of human mRNAs via post-transcriptional regulation. Interindividual variation of the miRNA expression levels is likely to influence the expression of miRNA target genes and may therefore contribute to phenotypic differences in humans, including susceptibility to common disorders. The extent to which miRNA levels are genetically controlled is largely unknown. In this report, we assayed the expression levels of miRNAs in primary fibroblasts from 180 European newborns of the GenCord project and performed association analysis to identify eQTLs (expression quantitative traits loci). We detected robust expression for 121 miRNAs out of 365 interrogated. We have identified significant cis- (10%) and trans- (11%) eQTLs. Furthermore, we detected one genomic locus (rs1522653) that influences the expression levels of five miRNAs, thus unraveling a novel mechanism for coregulation of miRNA expression.
"Intergenic transcription causes repression by directing nucleosome assembly [Research Papers]
Transcription of non-protein-coding DNA (ncDNA) and its noncoding RNA (ncRNA) products are beginning to emerge as key regulators of gene expression. We previously identified a regulatory system in Saccharomyces cerevisiae whereby transcription of intergenic ncDNA (SRG1) represses transcription of an adjacent protein-coding gene (SER3) through transcription interference. We now provide evidence that SRG1 transcription causes repression of SER3 by directing a high level of nucleosomes over SRG1, which overlaps the SER3 promoter. Repression by SRG1 transcription is dependent on the Spt6 and Spt16 transcription elongation factors. Significantly, spt6 and spt16 mutations reduce nucleosome levels over the SER3 promoter without reducing intergenic SRG1 transcription, strongly suggesting that nucleosome levels, not transcription levels, cause SER3 repression. Finally, we show that spt6 and spt16 mutations allow transcription factor access to the SER3 promoter. Our results raise the possibility that transcription of ncDNA may contribute to nucleosome positioning on a genome-wide scale where, in some cases, it negatively impacts protein–DNA interactions.
"