Wednesday, May 30, 2012
Diffusion in the early Drosophila embryo [Developmental Biology]
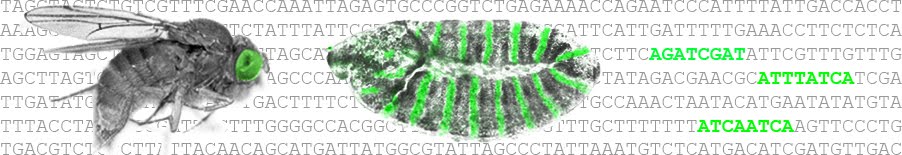
Diffusion in the early Drosophila embryo [Developmental Biology]: Despite the fundamental importance of diffusion for embryonic morphogen gradient formation in the early Drosophila melanogaster embryo, there remains controversy regarding both the extent and the rate of diffusion of well-characterized morphogens. Furthermore, the recent observation of diffusional “compartmentalization” has suggested that diffusion may in fact be nonideal and mediated by an as-yet-unidentified mechanism. Here, we characterize the effects of the geometry of the early syncytial Drosophila embryo on the effective diffusivity of cytoplasmic proteins. Our results demonstrate that the presence of transient mitotic membrane furrows results in a multiscale diffusion effect that has a significant impact on effective diffusion rates across the embryo. Using a combination of live-cell experiments and computational modeling, we characterize these effects and relate effective bulk diffusion rates to instantaneous diffusion coefficients throughout the syncytial blastoderm nuclear cycle phase of the early embryo. This multiscale effect may be related to the effect of interphase nuclei on effective diffusion, and thus we propose that an as-yet-unidentified role of syncytial membrane furrows is to temporally regulate bulk embryonic diffusion rates to balance the multiscale effect of interphase nuclei, which ultimately stabilizes the shapes of various morphogen gradients.
Tuesday, May 29, 2012
The adjustable nucleosome: an epigenetic signaling module
The adjustable nucleosome: an epigenetic signaling module: Bryan M. Turner. This review examines the proposition that the nucleosome, in addition to its role as a DNA packaging device, is a signaling module through which changing environmental and metabolic conditions can....
Tuesday, May 22, 2012
Origin of the beak-like dentition in pufferfish [Developmental Biology]
Origin of the beak-like dentition in pufferfish [Developmental Biology]: Teleost fishes comprise approximately half of all living vertebrates. The extreme range of diversity in teleosts is remarkable, especially, extensive morphological variation in their jaws and dentition. Some of the most unusual dentitions are found among members of the highly derived teleost order Tetraodontiformes, which includes triggerfishes, boxfishes, ocean sunfishes, and pufferfishes. Adult pufferfishes (Tetraodontidae) exhibit a distinctive parrot-like beaked jaw, forming a cutting edge, unlike in any other group of teleosts. Here we show that despite novelty in the structure and development of this “beak,” it is initiated by formation of separate first-generation teeth that line the embryonic pufferfish jaw, with timing of development and gene expression patterns conserved from the last common ancestor of osteichthyans. Most of these first-generation larval teeth are lost in development. Continuous tooth replacement proceeds in only four parasymphyseal teeth, as sequentially stacked, multigenerational, jaw-length dentine bands, before development of the functional beak. These data suggest that dental novelties, such as the pufferfish beak, can develop later in ontogeny through modified continuous tooth addition and replacement. We conclude that even highly derived morphological structures like the pufferfish beak form via a conserved developmental bauplan capable of modification during ontogeny by subtle respecification of the developmental module.
Evolution of a tissue-specific silencer underlies divergence in the expression of pax2 and pax8 paralogues
Evolution of a tissue-specific silencer underlies divergence in the expression of pax2 and pax8 paralogues:
Evolution of a tissue-specific silencer underlies divergence in the expression of pax2 and pax8 paralogues
Nature Communications 3, 848 (2012). doi:10.1038/ncomms1851
Authors: Haruki Ochi, Tomoko Tamai, Hiroki Nagano, Akane Kawaguchi, Norihiro Sudou & Hajime Ogino

Evolution of a tissue-specific silencer underlies divergence in the expression of pax2 and pax8 paralogues
Nature Communications 3, 848 (2012). doi:10.1038/ncomms1851
Authors: Haruki Ochi, Tomoko Tamai, Hiroki Nagano, Akane Kawaguchi, Norihiro Sudou & Hajime Ogino
Friday, May 18, 2012
Transcriptional Dominance of Pax7 in Adult Myogenesis Is Due to High-Affinity Recognition of Homeodomain Motifs
Transcriptional Dominance of Pax7 in Adult Myogenesis Is Due to High-Affinity Recognition of Homeodomain Motifs: Vahab D. Soleimani, Vincent G. Punch, Yoh-ichi Kawabe, Andrew E. Jones, Gareth A. Palidwor, Christopher J. Porter, Joe W. Cross, Jaime J. Carvajal, Christel E.M. Kockx, Wilfred F.J. van IJcken, Theodore J. Perkins, Peter W.J. Rigby, Frank Grosveld, Michael A. Rudnicki. Pax3 and Pax7 regulate stem cell function in skeletal myogenesis. However, molecular insight into their distinct roles has remained elusive. Using gene expression data combined with genome-wide bi....
Wednesday, May 16, 2012
Topological domains in mammalian genomes identified by analysis of chromatin interactions
Topological domains in mammalian genomes identified by analysis of chromatin interactions:
Topological domains in mammalian genomes identified by analysis of chromatin interactions
Nature 485, 7398 (2012). doi:10.1038/nature11082
Authors: Jesse R. Dixon, Siddarth Selvaraj, Feng Yue, Audrey Kim, Yan Li, Yin Shen, Ming Hu, Jun S. Liu & Bing Ren
The spatial organization of the genome is intimately linked to its biological function, yet our understanding of higher order genomic structure is coarse, fragmented and incomplete. In the nucleus of eukaryotic cells, interphase chromosomes occupy distinct chromosome territories, and numerous models have been proposed for how chromosomes fold within chromosome territories. These models, however, provide only few mechanistic details about the relationship between higher order chromatin structure and genome function. Recent advances in genomic technologies have led to rapid advances in the study of three-dimensional genome organization. In particular, Hi-C has been introduced as a method for identifying higher order chromatin interactions genome wide. Here we investigate the three-dimensional organization of the human and mouse genomes in embryonic stem cells and terminally differentiated cell types at unprecedented resolution. We identify large, megabase-sized local chromatin interaction domains, which we term ‘topological domains’, as a pervasive structural feature of the genome organization. These domains correlate with regions of the genome that constrain the spread of heterochromatin. The domains are stable across different cell types and highly conserved across species, indicating that topological domains are an inherent property of mammalian genomes. Finally, we find that the boundaries of topological domains are enriched for the insulator binding protein CTCF, housekeeping genes, transfer RNAs and short interspersed element (SINE) retrotransposons, indicating that these factors may have a role in establishing the topological domain structure of the genome.

Topological domains in mammalian genomes identified by analysis of chromatin interactions
Nature 485, 7398 (2012). doi:10.1038/nature11082
Authors: Jesse R. Dixon, Siddarth Selvaraj, Feng Yue, Audrey Kim, Yan Li, Yin Shen, Ming Hu, Jun S. Liu & Bing Ren
The spatial organization of the genome is intimately linked to its biological function, yet our understanding of higher order genomic structure is coarse, fragmented and incomplete. In the nucleus of eukaryotic cells, interphase chromosomes occupy distinct chromosome territories, and numerous models have been proposed for how chromosomes fold within chromosome territories. These models, however, provide only few mechanistic details about the relationship between higher order chromatin structure and genome function. Recent advances in genomic technologies have led to rapid advances in the study of three-dimensional genome organization. In particular, Hi-C has been introduced as a method for identifying higher order chromatin interactions genome wide. Here we investigate the three-dimensional organization of the human and mouse genomes in embryonic stem cells and terminally differentiated cell types at unprecedented resolution. We identify large, megabase-sized local chromatin interaction domains, which we term ‘topological domains’, as a pervasive structural feature of the genome organization. These domains correlate with regions of the genome that constrain the spread of heterochromatin. The domains are stable across different cell types and highly conserved across species, indicating that topological domains are an inherent property of mammalian genomes. Finally, we find that the boundaries of topological domains are enriched for the insulator binding protein CTCF, housekeeping genes, transfer RNAs and short interspersed element (SINE) retrotransposons, indicating that these factors may have a role in establishing the topological domain structure of the genome.
Tuesday, May 15, 2012
Orchestrating transcriptional control of adult neurogenesis [Reviews]
Orchestrating transcriptional control of adult neurogenesis [Reviews]:
Stem cells have captured our imagination and generated hope, representing a source of replacement cells to treat a host of medical conditions. Tucked away in specialized niches, stem cells maintain tissue function and rejuvenate organs. Balancing the equation between cellular supply and demand is especially important in the adult brain, as neural stem cells (NSCs) in two discrete regions, the subgranular zone (SGZ) of the dentate gyrus and the subventricular zone (SVZ) next to the lateral ventricles, continuously self-renew and differentiate into neurons in a process called adult neurogenesis. Through the interplay of intrinsic and extrinsic factors, adult neurogenic niches ensure neuronal turnover throughout life, contributing to plasticity and homeostatic processes in the brain. This review summarizes recent progress on the molecular control of adult neurogenesis in the SGZ and SVZ, focusing on the role of specific transcription factors that mediate the progression from NSCs to lineage-committed progenitors and, ultimately, the generation of mature neurons and glia.
Stem cells have captured our imagination and generated hope, representing a source of replacement cells to treat a host of medical conditions. Tucked away in specialized niches, stem cells maintain tissue function and rejuvenate organs. Balancing the equation between cellular supply and demand is especially important in the adult brain, as neural stem cells (NSCs) in two discrete regions, the subgranular zone (SGZ) of the dentate gyrus and the subventricular zone (SVZ) next to the lateral ventricles, continuously self-renew and differentiate into neurons in a process called adult neurogenesis. Through the interplay of intrinsic and extrinsic factors, adult neurogenic niches ensure neuronal turnover throughout life, contributing to plasticity and homeostatic processes in the brain. This review summarizes recent progress on the molecular control of adult neurogenesis in the SGZ and SVZ, focusing on the role of specific transcription factors that mediate the progression from NSCs to lineage-committed progenitors and, ultimately, the generation of mature neurons and glia.
Monday, May 14, 2012
Transcription Factor Binding to a DNA Zip Code Controls Interchromosomal Clustering at the Nuclear Periphery
Transcription Factor Binding to a DNA Zip Code Controls Interchromosomal Clustering at the Nuclear Periphery: Donna Garvey Brickner, Sara Ahmed, Lauren Meldi, Abbey Thompson, Will Light, Matthew Young, Taylor L. Hickman, Feixia Chu, Emmanuelle Fabre, Jason H. Brickner. Active genes in yeast can be targeted to the nuclear periphery through interaction of cis-acting “DNA zip codes” with the nuclear pore complex. We find that genes with identical zip codes c....
Thursday, May 10, 2012
Identification and Characterization of Lineage-Specific Highly Conserved Noncoding Sequences in Mammalian Genomes
Identification and Characterization of Lineage-Specific Highly Conserved Noncoding Sequences in Mammalian Genomes:
Vertebrate genome comparisons revealed that there are highly conserved noncoding sequences (HCNSs) among a wide range of species and many of which contain regulatory elements. However, recently emerged sequences conserved in specific lineages have not been well studied. Toward this end, we identified 8,198 primate and 21,128 specific HCNSs as representative ones among mammals from human–marmoset and mouse–rat comparisons, respectively. Derived allele frequency analysis of primate-specific HCNSs showed that these HCNSs were under purifying selection, indicating that they may harbor important functions. We selected the top 1,000 largest HCNSs and compared the lineage-specific HCNS-flanking genes (LHF genes) with ultraconserved element (UCE)-flanking genes. Interestingly, the majority of LHF genes were different from UCE-flanking genes. This lineage-specific set of LHF genes was more enriched in protein-binding function. Conversely, the number of LHF genes that were also shared by UCEs was small but significantly larger than random expectation, and many of these genes were involved in anatomical development as transcriptional regulators, suggesting that certain groups of genes preferentially recruit new HCNSs in addition to old HCNSs that are conserved among vertebrates. This group of LHF genes might be involved in the various levels of lineage-specific evolution among vertebrates, mammals, primates, and rodents. If so, the emergence of HCNSs in and around these two groups of LHF genes developed lineage-specific characteristics. Our results provide new insight into lineage-specific evolution through interactions between HCNSs and their LHF genes.
Vertebrate genome comparisons revealed that there are highly conserved noncoding sequences (HCNSs) among a wide range of species and many of which contain regulatory elements. However, recently emerged sequences conserved in specific lineages have not been well studied. Toward this end, we identified 8,198 primate and 21,128 specific HCNSs as representative ones among mammals from human–marmoset and mouse–rat comparisons, respectively. Derived allele frequency analysis of primate-specific HCNSs showed that these HCNSs were under purifying selection, indicating that they may harbor important functions. We selected the top 1,000 largest HCNSs and compared the lineage-specific HCNS-flanking genes (LHF genes) with ultraconserved element (UCE)-flanking genes. Interestingly, the majority of LHF genes were different from UCE-flanking genes. This lineage-specific set of LHF genes was more enriched in protein-binding function. Conversely, the number of LHF genes that were also shared by UCEs was small but significantly larger than random expectation, and many of these genes were involved in anatomical development as transcriptional regulators, suggesting that certain groups of genes preferentially recruit new HCNSs in addition to old HCNSs that are conserved among vertebrates. This group of LHF genes might be involved in the various levels of lineage-specific evolution among vertebrates, mammals, primates, and rodents. If so, the emergence of HCNSs in and around these two groups of LHF genes developed lineage-specific characteristics. Our results provide new insight into lineage-specific evolution through interactions between HCNSs and their LHF genes.
Monday, May 7, 2012
Tuesday, May 1, 2012
HOT regions function as patterned developmental enhancers and have a distinct cis-regulatory signature [Research Communications]
HOT regions function as patterned developmental enhancers and have a distinct cis-regulatory signature [Research Communications]:
HOT (highly occupied target) regions bound by many transcription factors are considered to be one of the most intriguing findings of the recent modENCODE reports, yet their functions have remained unclear. We tested 108 Drosophila melanogaster HOT regions in transgenic embryos with site-specifically integrated transcriptional reporters. In contrast to prior expectations, we found 102 (94%) to be active enhancers during embryogenesis and to display diverse spatial and temporal patterns, reminiscent of expression patterns for important developmental genes. Remarkably, HOT regions strongly activate nearby genes and are required for endogenous gene expression, as we show using bacterial artificial chromosome (BAC) transgenesis. HOT enhancers have a distinct cis-regulatory signature with enriched sequence motifs for the global activators Vielfaltig, also known as Zelda, and Trithorax-like, also known as GAGA. This signature allows the prediction of HOT versus control regions from the DNA sequence alone.
HOT (highly occupied target) regions bound by many transcription factors are considered to be one of the most intriguing findings of the recent modENCODE reports, yet their functions have remained unclear. We tested 108 Drosophila melanogaster HOT regions in transgenic embryos with site-specifically integrated transcriptional reporters. In contrast to prior expectations, we found 102 (94%) to be active enhancers during embryogenesis and to display diverse spatial and temporal patterns, reminiscent of expression patterns for important developmental genes. Remarkably, HOT regions strongly activate nearby genes and are required for endogenous gene expression, as we show using bacterial artificial chromosome (BAC) transgenesis. HOT enhancers have a distinct cis-regulatory signature with enriched sequence motifs for the global activators Vielfaltig, also known as Zelda, and Trithorax-like, also known as GAGA. This signature allows the prediction of HOT versus control regions from the DNA sequence alone.
Effects of sequence variation on differential allelic transcription factor occupancy and gene expression [RESEARCH]
Effects of sequence variation on differential allelic transcription factor occupancy and gene expression [RESEARCH]:
A complex interplay between transcription factors (TFs) and the genome regulates transcription. However, connecting variation in genome sequence with variation in TF binding and gene expression is challenging due to environmental differences between individuals and cell types. To address this problem, we measured genome-wide differential allelic occupancy of 24 TFs and EP300 in a human lymphoblastoid cell line GM12878. Overall, 5% of human TF binding sites have an allelic imbalance in occupancy. At many sites, TFs clustered in TF-binding hubs on the same homolog in especially open chromatin. While genetic variation in core TF binding motifs generally resulted in large allelic differences in TF occupancy, most allelic differences in occupancy were subtle and associated with disruption of weak or noncanonical motifs. We also measured genome-wide differential allelic expression of genes with and without heterozygous exonic variants in the same cells. We found that genes with differential allelic expression were overall less expressed both in GM12878 cells and in unrelated human cell lines. Comparing TF occupancy with expression, we found strong association between allelic occupancy and expression within 100 bp of transcription start sites (TSSs), and weak association up to 100 kb from TSSs. Sites of differential allelic occupancy were significantly enriched for variants associated with disease, particularly autoimmune disease, suggesting that allelic differences in TF occupancy give functional insights into intergenic variants associated with disease. Our results have the potential to increase the power and interpretability of association studies by targeting functional intergenic variants in addition to protein coding sequences.
A complex interplay between transcription factors (TFs) and the genome regulates transcription. However, connecting variation in genome sequence with variation in TF binding and gene expression is challenging due to environmental differences between individuals and cell types. To address this problem, we measured genome-wide differential allelic occupancy of 24 TFs and EP300 in a human lymphoblastoid cell line GM12878. Overall, 5% of human TF binding sites have an allelic imbalance in occupancy. At many sites, TFs clustered in TF-binding hubs on the same homolog in especially open chromatin. While genetic variation in core TF binding motifs generally resulted in large allelic differences in TF occupancy, most allelic differences in occupancy were subtle and associated with disruption of weak or noncanonical motifs. We also measured genome-wide differential allelic expression of genes with and without heterozygous exonic variants in the same cells. We found that genes with differential allelic expression were overall less expressed both in GM12878 cells and in unrelated human cell lines. Comparing TF occupancy with expression, we found strong association between allelic occupancy and expression within 100 bp of transcription start sites (TSSs), and weak association up to 100 kb from TSSs. Sites of differential allelic occupancy were significantly enriched for variants associated with disease, particularly autoimmune disease, suggesting that allelic differences in TF occupancy give functional insights into intergenic variants associated with disease. Our results have the potential to increase the power and interpretability of association studies by targeting functional intergenic variants in addition to protein coding sequences.
Subscribe to:
Posts (Atom)