Tuesday, January 31, 2012
Experimental evolution of multicellularity [Evolution]
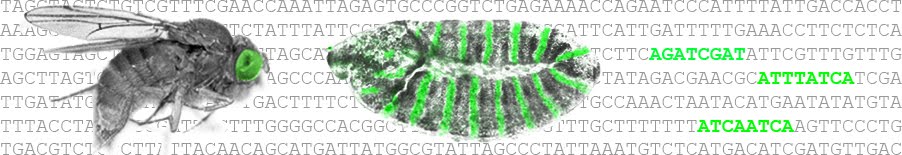
Transcriptional interpretation of the EGF receptor signaling gradient [Developmental Biology]
Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development
Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development
Nature Genetics 44, 148 (2012).
doi:10.1038/ng.1064
Authors: Stefan Bonn, Robert P Zinzen, Charles Girardot, E Hilary Gustafson, Alexis Perez-Gonzalez, Nicolas Delhomme, Yad Ghavi-Helm, Bartek Wilczyński, Andrew Riddell & Eileen E M Furlong
Thursday, January 26, 2012
Sex Pheromone Evolution Is Associated with Differential Regulation of the Same Desaturase Gene in Two Genera of Leafroller Moths
by Jérôme Albre, Marjorie A. Liénard, Tamara M. Sirey, Silvia Schmidt, Leah K. Tooman, Colm Carraher, David R. Greenwood, Christer Löfstedt, Richard D. Newcomb
Chemical signals are prevalent in sexual communication systems. Mate recognition has been extensively studied within the Lepidoptera, where the production and recognition of species-specific sex pheromone signals are typically the defining character. While the specific blend of compounds that makes up the sex pheromones of many species has been characterized, the molecular mechanisms underpinning the evolution of pheromone-based mate recognition systems remain largely unknown. We have focused on two sets of sibling species within the leafroller moth genera Ctenopseustis and Planotortrix that have rapidly evolved the use of distinct sex pheromone blends. The compounds within these blends differ almost exclusively in the relative position of double bonds that are introduced by desaturase enzymes. Of the six desaturase orthologs isolated from all four species, functional analyses in yeast and gene expression in pheromone glands implicate three in pheromone biosynthesis, two Δ9-desaturases, and a Δ10-desaturase, while the remaining three desaturases include a Δ6-desaturase, a terminal desaturase, and a non-functional desaturase. Comparative quantitative real-time PCR reveals that the Δ10-desaturase is differentially expressed in the pheromone glands of the two sets of sibling species, consistent with differences in the pheromone blend in both species pairs. In the pheromone glands of species that utilize (Z)-8-tetradecenyl acetate as sex pheromone component (Ctenopseustis obliquana and Planotortrix octo), the expression levels of the Δ10-desaturase are significantly higher than in the pheromone glands of their respective sibling species (C. herana and P. excessana). Our results demonstrate that interspecific sex pheromone differences are associated with differential regulation of the same desaturase gene in two genera of moths. We suggest that differential gene regulation among members of a multigene family may be an important mechanism of molecular innovation in sex pheromone evolution and speciation.
Wednesday, January 25, 2012
Stochastic Expression of the Interferon-β Gene
by Mingwei Zhao, Jiangwen Zhang, Hemali Phatnani, Stefanie Scheu, Tom Maniatis
Virus infection of mammalian cells induces the production of high levels of type I interferons (IFNα and β), cytokines that orchestrate antiviral innate and adaptive immunity. Previous studies have shown that only a fraction of the infected cells produce IFN. However, the mechanisms responsible for this stochastic expression are poorly understood. Here we report an in depth analysis of IFN-expressing and non-expressing mouse cells infected with Sendai virus. Mouse embryonic fibroblasts in which an internal ribosome entry site/yellow fluorescent protein gene was inserted downstream from the endogenous IFNβ gene were used to distinguish between the two cell types, and they were isolated from each other using fluorescence-activated cell sorting methods. Analysis of the separated cells revealed that stochastic IFNβ expression is a consequence of cell-to-cell variability in the levels and/or activities of limiting components at every level of the virus induction process, ranging from viral replication and expression, to the sensing of viral RNA by host factors, to activation of the signaling pathway, to the levels of activated transcription factors. We propose that this highly complex stochastic IFNβ gene expression evolved to optimize both the level and distribution of type I IFNs in response to virus infection.
Friday, January 20, 2012
Gene Regulatory Logic for Reading the Sonic Hedgehog Signaling Gradient in the Vertebrate Neural Tube
Extensive Promoter-Centered Chromatin Interactions Provide a Topological Basis for Transcription Regulation
Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome
Thursday, January 19, 2012
Tempo and Mode in Evolution of Transcriptional Regulation
by Kacy L. Gordon, Ilya Ruvinsky
Perennial questions of evolutionary biology can be applied to gene regulatory systems using the abundance of experimental data addressing gene regulation in a comparative context. What is the tempo (frequency, rate) and mode (way, mechanism) of transcriptional regulatory evolution? Here we synthesize the results of 230 experiments performed on insects and nematodes in which regulatory DNA from one species was used to drive gene expression in another species. General principles of regulatory evolution emerge. Gene regulatory evolution is widespread and accumulates with genetic divergence in both insects and nematodes. Divergence in cis is more common than divergence in trans. Coevolution between cis and trans shows a particular increase over greater evolutionary timespans, especially in sex-specific gene regulation. Despite these generalities, the evolution of gene regulation is gene- and taxon-specific. The congruence of these conclusions with evidence from other types of experiments suggests that general principles are discoverable, and a unified view of the tempo and mode of regulatory evolution may be achievable.
Unraveling the Regulatory Mechanisms Underlying Tissue-Dependent Genetic Variation of Gene Expression
by Jingyuan Fu, Marcel G. M. Wolfs, Patrick Deelen, Harm-Jan Westra, Rudolf S. N. Fehrmann, Gerard J. te Meerman, Wim A. Buurman, Sander S. M. Rensen, Harry J. M. Groen, Rinse K. Weersma, Leonard H. van den Berg, Jan Veldink, Roel A. Ophoff, Harold Snieder, David van Heel, Ritsert C. Jansen, Marten H. Hofker, Cisca Wijmenga, Lude Franke
It is known that genetic variants can affect gene expression, but it is not yet completely clear through what mechanisms genetic variation mediate this expression. We therefore compared the cis-effect of single nucleotide polymorphisms (SNPs) on gene expression between blood samples from 1,240 human subjects and four primary non-blood tissues (liver, subcutaneous, and visceral adipose tissue and skeletal muscle) from 85 subjects. We characterized four different mechanisms for 2,072 probes that show tissue-dependent genetic regulation between blood and non-blood tissues: on average 33.2% only showed cis-regulation in non-blood tissues; 14.5% of the eQTL probes were regulated by different, independent SNPs depending on the tissue of investigation. 47.9% showed a different effect size although they were regulated by the same SNPs. Surprisingly, we observed that 4.4% were regulated by the same SNP but with opposite allelic direction. We show here that SNPs that are located in transcriptional regulatory elements are enriched for tissue-dependent regulation, including SNPs at 3′ and 5′ untranslated regions (P = 1.84×10−5 and 4.7×10−4, respectively) and SNPs that are synonymous-coding (P = 9.9×10−4). SNPs that are associated with complex traits more often exert a tissue-dependent effect on gene expression (P = 2.6×10−10). Our study yields new insights into the genetic basis of tissue-dependent expression and suggests that complex trait associated genetic variants have even more complex regulatory effects than previously anticipated.
Wednesday, January 18, 2012
Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription
Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription
Nature Reviews Genetics 13, 123 (2012).
doi:10.1038/nrg3124
Authors: Thomas Conrad & Asifa Akhtar
Dosage compensation is an epigenetic mechanism that normalizes gene expression from unequal copy numbers of sex chromosomes. Different organisms have evolved alternative molecular solutions to this task. In Drosophila melanogaster, transcription of the single male X chromosome is upregulated by twofold in a process
Friday, January 13, 2012
Evolution at the Subgene Level: Domain Rearrangements in the Drosophila Phylogeny
Although the possibility of gene evolution by domain rearrangements has long been appreciated, current methods for reconstructing and systematically analyzing gene family evolution are limited to events such as duplication, loss, and sometimes, horizontal transfer. However, within the Drosophila clade, we find domain rearrangements occur in 35.9% of gene families, and thus, any comprehensive study of gene evolution in these species will need to account for such events. Here, we present a new computational model and algorithm for reconstructing gene evolution at the domain level. We develop a method for detecting homologous domains between genes and present a phylogenetic algorithm for reconstructing maximum parsimony evolutionary histories that include domain generation, duplication, loss, merge (fusion), and split (fission) events. Using this method, we find that genes involved in fusion and fission are enriched in signaling and development, suggesting that domain rearrangements and reuse may be crucial in these processes. We also find that fusion is more abundant than fission, and that fusion and fission events occur predominantly alongside duplication, with 92.5% and 34.3% of fusion and fission events retaining ancestral architectures in the duplicated copies. We provide a catalog of ~9,000 genes that undergo domain rearrangement across nine sequenced species, along with possible mechanisms for their formation. These results dramatically expand on evolution at the subgene level and offer several insights into how new genes and functions arise between species.
Chimeric Genes as a Source of Rapid Evolution in Drosophila melanogaster
Chimeric genes form through the combination of portions of existing coding sequences to create a new open reading frame. These new genes can create novel protein structures that are likely to serve as a strong source of novelty upon which selection can act. We have identified 14 chimeric genes that formed through DNA-level mutations in Drosophila melanogaster, and we investigate expression profiles, domain structures, and population genetics for each of these genes to examine their potential to effect adaptive evolution. We find that chimeric gene formation commonly produces mid-domain breaks and unites portions of wholly unrelated peptides, creating novel protein structures that are entirely distinct from other constructs in the genome. These new genes are often involved in selective sweeps. We further find a disparity between chimeric genes that have recently formed and swept to fixation versus chimeric genes that have been preserved over long periods of time, suggesting that preservation and adaptation are distinct processes. Finally, we demonstrate that chimeric gene formation can produce qualitative expression changes that are difficult to mimic through duplicate gene formation, and that extremely young chimeric genes (dS < 0.03) are more likely to be associated with selective sweeps than duplicate genes of the same age. Hence, chimeric genes can serve as an exceptional source of genetic novelty that can have a profound influence on adaptive evolution in D. melanogaster.
Wednesday, January 11, 2012
Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle [Genetics]
Wednesday, January 4, 2012
Role of Pleiotropy in the Evolution of a Cryptic Developmental Variation in Caenorhabditis elegans
by Fabien Duveau, Marie-Anne Félix
Robust biological systems are expected to accumulate cryptic genetic variation that does not affect the system output in standard conditions yet may play an evolutionary role once phenotypically expressed under a strong perturbation. Genetic variation that is cryptic relative to a robust trait may accumulate neutrally as it does not change the phenotype, yet it could also evolve under selection if it affects traits related to fitness in addition to its cryptic effect. Cryptic variation affecting the vulval intercellular signaling network was previously uncovered among wild isolates of Caenorhabditis elegans. Using a quantitative genetic approach, we identify a non-synonymous polymorphism of the previously uncharacterized nath-10 gene that affects the vulval phenotype when the system is sensitized with different mutations, but not in wild-type strains. nath-10 is an essential protein acetyltransferase gene and the homolog of human NAT10. The nath-10 polymorphism also presents non-cryptic effects on life history traits. The nath-10 allele carried by the N2 reference strain leads to a subtle increase in the egg laying rate and in the total number of sperm, a trait affecting the trade-off between fertility and minimal generation time in hermaphrodite individuals. We show that this allele appeared during early laboratory culture of N2, which allowed us to test whether it may have evolved under selection in this novel environment. The derived allele indeed strongly outcompetes the ancestral allele in laboratory conditions. In conclusion, we identified the molecular nature of a cryptic genetic variation and characterized its evolutionary history. These results show that cryptic genetic variation does not necessarily accumulate neutrally at the whole-organism level, but may evolve through selection for pleiotropic effects that alter fitness. In addition, cultivation in the laboratory has led to adaptive evolution of the reference strain N2 to the laboratory environment, which may modify other phenotypes of interest.
Expression of a desaturase gene, desat1, in neural and nonneural tissues separately affects perception and emission of sex pheromones in Drosophila [Genetics]
Genetic adaptation to captivity can occur in a single generation [Evolution]
Transposase mediated construction of RNA-seq libraries [METHOD]
RNA-seq has been widely adopted as a gene-expression measurement tool due to the detail, resolution, and sensitivity of transcript characterization that the technique provides. Here we present two transposon-based methods that efficiently construct high-quality RNA-seq libraries. We first describe a method that creates RNA-seq libraries for Illumina sequencing from double-stranded cDNA with only two enzymatic reactions. We generated high-quality RNA-seq libraries from as little as 10 pg of mRNA (~1 ng of total RNA) with this approach. We also present a strand-specific RNA-seq library construction protocol that combines transposon-based library construction with uracil DNA glycosylase and endonuclease VIII to specifically degrade the second strand constructed during cDNA synthesis. The directional RNA-seq libraries maintain the same quality as the nondirectional libraries, while showing a high degree of strand specificity, such that 99.5% of reads map to the expected genomic strand. Each transposon-based library construction method performed well when compared with standard RNA-seq library construction methods with regard to complexity of the libraries, correlation between biological replicates, and the percentage of reads that align to the genome as well as exons. Our results show that high-quality RNA-seq libraries can be constructed efficiently and in an automatable fashion using transposition technology.
A novel candidate cis-regulatory motif pair in the promoters of germline and oogenesis genes in C. elegans [RESEARCH]
In this study we report on a novel pair of cis-regulatory motifs in promoter sequences of the nematode Caenorhabditis elegans. The motif pair exhibits extraordinary genomic traits: The order and the orientation of the two motifs are highly specific, and the distance between them is almost always one of two frequent distances. In contrast, the sequence between the motifs is variable across occurrences. Thus, the motif pair constitutes a nearly combinatorial sequence configuration. We further show that this module is conserved among, and unique to, the entire Caenorhabditis genus. By analyzing several gene expression data sets, our data suggest that this motif pair may function in germline development, oogenesis, and early embryogenesis. Finally, we verify that the motifs are indeed functional cis-regulatory elements using reporter constructs in transgenic C. elegans.
Cell-type specific and combinatorial usage of diverse transcription factors revealed by genome-wide binding studies in multiple human cells [RESEARCH]
Cell-type diversity is governed in part by differential gene expression programs mediated by transcription factor (TF) binding. However, there are few systematic studies of the genomic binding of different types of TFs across a wide range of human cell types, especially in relation to gene expression. In the ENCODE Project, we have identified the genomic binding locations across 11 different human cell types of CTCF, RNA Pol II (RNAPII), and MYC, three TFs with diverse roles. Our data and analysis revealed how these factors bind in relation to genomic features and shape gene expression and cell-type specificity. CTCF bound predominantly in intergenic regions while RNAPII and MYC preferentially bound to core promoter regions. CTCF sites were relatively invariant across diverse cell types, while MYC showed the greatest cell-type specificity. MYC and RNAPII co-localized at many of their binding sites and putative target genes. Cell-type specific binding sites, in particular for MYC and RNAPII, were associated with cell-type specific functions. Patterns of binding in relation to gene features were generally conserved across different cell types. RNAPII occupancy was higher over exons than adjacent introns, likely reflecting a link between transcriptional elongation and splicing. TF binding was positively correlated with the expression levels of their putative target genes, but combinatorial binding, in particular of MYC and RNAPII, was even more strongly associated with higher gene expression. These data illuminate how combinatorial binding of transcription factors in diverse cell types is associated with gene expression and cell-type specific biology.
A decade of 3C technologies: insights into nuclear organization [Reviews]
Over the past 10 years, the development of chromosome conformation capture (3C) technology and the subsequent genomic variants thereof have enabled the analysis of nuclear organization at an unprecedented resolution and throughput. The technology relies on the original and, in hindsight, remarkably simple idea that digestion and religation of fixed chromatin in cells, followed by the quantification of ligation junctions, allows for the determination of DNA contact frequencies and insight into chromosome topology. Here we evaluate and compare the current 3C-based methods (including 4C [chromosome conformation capture-on-chip], 5C [chromosome conformation capture carbon copy], HiC, and ChIA-PET), summarize their contribution to our current understanding of genome structure, and discuss how shape influences genome function.
Sunday, January 1, 2012
Ancient Pbx-Hox Signatures Define Hundreds of Vertebrate Developmental Enhancers
Gene regulation through cis-regulatory elements plays a crucial role in development and disease. A major aim of the post-genomic era is to be able to read the function of cis-regulatory elements through scrutiny of their DNA sequence. Whilst comparative genomics approaches have identified thousands of putative regulatory elements, our knowledge of their mechanism of action is poor and very little progress has been made in systematically de-coding them.
Results:
Here, we identify ancient functional signatures within vertebrate conserved non-coding elements (CNEs) through a combination of phylogenetic footprinting and functional assay, using genomic sequence from the sea lamprey as a reference. We uncover a striking enrichment within vertebrate CNEs for conserved binding-site motifs of the Pbx-Hox hetero-dimer. We further show that these predict reporter gene expression in a segment specific manner in the hindbrain and pharyngeal arches during zebrafish development.
Conclusions:
These findings evoke an evolutionary scenario in which many CNEs evolved early in the vertebrate lineage to co-ordinate Hox-dependent gene-regulatory interactions that pattern the vertebrate head. In a broader context, our evolutionary analyses reveal that CNEs are composed of tightly linked transcription-factor binding-sites (TFBSs), which can be systematically identified through phylogenetic footprinting approaches. By placing a large number of ancient vertebrate CNEs into a developmental context, our findings promise to have a significant impact on efforts toward de-coding gene-regulatory elements that underlie vertebrate development, and will facilitate building general models of regulatory element evolution.