A unique chromatin signature uncovers early developmental enhancers in humans: "
A unique chromatin signature uncovers early developmental enhancers in humans
Nature 470, 7333 (2011). doi:10.1038/nature09692
Authors: Alvaro Rada-Iglesias, Ruchi Bajpai, Tomek Swigut, Samantha A. Brugmann, Ryan A. Flynn & Joanna Wysocka
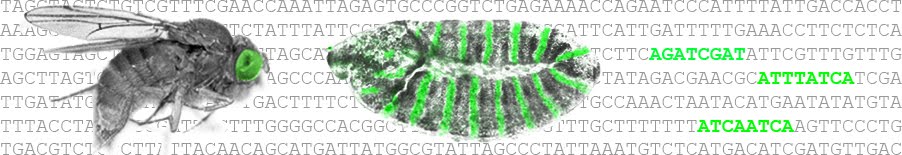
Cell-fate transitions involve the integration of genomic information encoded by regulatory elements, such as enhancers, with the cellular environment. However, identification of genomic sequences that control human embryonic development represents a formidable challenge. Here we show that in human embryonic stem cells (hESCs), unique chromatin signatures identify two distinct classes of genomic elements, both of which are marked by the presence of chromatin regulators p300 and BRG1, monomethylation of histone H3 at lysine 4 (H3K4me1), and low nucleosomal density. In addition, elements of the first class are distinguished by the acetylation of histone H3 at lysine 27 (H3K27ac), overlap with previously characterized hESC enhancers, and are located proximally to genes expressed in hESCs and the epiblast. In contrast, elements of the second class, which we term ‘poised enhancers’, are distinguished by the absence of H3K27ac, enrichment of histone H3 lysine 27 trimethylation (H3K27me3), and are linked to genes inactive in hESCs and instead are involved in orchestrating early steps in embryogenesis, such as gastrulation, mesoderm formation and neurulation. Consistent with the poised identity, during differentiation of hESCs to neuroepithelium, a neuroectoderm-specific subset of poised enhancers acquires a chromatin signature associated with active enhancers. When assayed in zebrafish embryos, poised enhancers are able to direct cell-type and stage-specific expression characteristic of their proximal developmental gene, even in the absence of sequence conservation in the fish genome. Our data demonstrate that early developmental enhancers are epigenetically pre-marked in hESCs and indicate an unappreciated role of H3K27me3 at distal regulatory elements. Moreover, the wealth of new regulatory sequences identified here provides an invaluable resource for studies and isolation of transient, rare cell populations representing early stages of human embryogenesis.

"